by Julian Göltz (Heidelberg University, University of Bern), Laura Kriener (University of Bern), Virginie Sabado (University of Bern) and Mihai A. Petrovici (University of Bern, Heidelberg University)
Many neuromorphic platforms promise fast and energy-efficient emulation of spiking neural networks, but unlike artificial neural networks, spiking networks have lacked a powerful universal training algorithm for more challenging machine learning applications. Such a training scheme has recently been proposed and using it together with a biologically inspired form of information coding shows state-of-the-art results in terms of classification accuracy, speed and energy consumption.
Spikes are the fundamental unit in which information is processed in mammalian brains, and a significant part of the information is encoded in the relative timing of these spikes. In contrast, the computational units of typical machine learning models output a numeric value without an accompanying time. This observation is, in a modified form, at the centre of a new approach: a network model and learning algorithm that can efficiently solve pattern recognition problems by making full use of the timing of spikes [1]. This quintessential reliance on spike-based communication perfectly synergises with efficient neuromorphic spiking-network emulators, such as the BrainScaleS-2 platform [2], thus being able to fully harness their speed and energy characteristics. This work is the result of a collaboration between neuromorphic engineers at the Heidelberg University and computational neuroscientists at the University of Bern, fostered by the European Human Brain Project.
In the implementation on BrainScaleS-2, to further enforce fast computation and to minimise resource requirements, an encoding was chosen where more prominent features are represented by earlier spikes, as seen in nature, e.g., in how nerves in the fingertips encode information about touch (Figure 1). From the perspective of an animal looking to survive, this choice of coding is particularly appealing, as actions must often be taken under time pressure. The biological imperative of short times-to-solution is similarly applicable to silicon, carrying with it an optimised usage of resources. In this model of neural computation, synaptic plasticity implements a version of error backpropagation on first-spike times, which we discuss in more detail below.
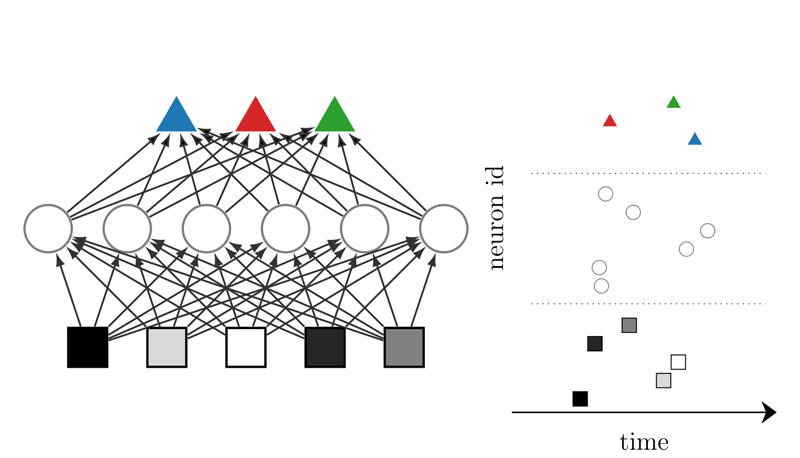
Figure 1: (Left) Discriminator network consisting of neurons (squares, circles, and triangles) grouped in layers. Information is passed from the bottom to the top, e.g. pixel brightness of an image. Here, a darker pixel is represented by an earlier spike. (Right) Each neuron spikes no more than once, and the time at which it spikes encodes the information.
This algorithm was demonstrated on the BrainScaleS-2 neuromorphic platform using both an artificial dataset resembling the Yin-Yang symbol [3], as well as the real-world MNIST data set of handwritten digits. The Yin-Yang dataset highlights the universality of the algorithm and its interplay with first-spike coding in a small network, ensuring that training of this classification problem achieves highly accurate results (Figure 2). For the digit-recognition problem, an optimised implementation yields particularly compelling results: up to 10,000 images can be classified in less than a second at a runtime power consumption of only 270 mW, which translates to only 27 µJ per image. For comparison, the power drawn by the BrainScaleS-2 chip for this application is about the same as a few LEDs.
![Figure 2: Artificial dataset resembling the Yin Yang symbol [3], and the output of a network trained with our algorithm. The goal of each of the target neurons is to spike as early as possible (small delay, bright yellow color) when a data point, represented as a circle, is “in their area”. One can see that the three neurons cover the correct areas in bright yellow.](/images/stories/EN125/goeltz2.png)
Figure 2: Artificial dataset resembling the Yin Yang symbol [3], and the output of a network trained with our algorithm. The goal of each of the target neurons is to spike as early as possible (small delay, bright yellow color) when a data point, represented as a circle, is “in their area”. One can see that the three neurons cover the correct areas in bright yellow.
![Figure 3: Comparison with other spike-based neuromorphic classifiers on the MNIST data set, see [1] for details. [A]: E. Stromatias et al. 2015, [B]: S. Esser et al. 2015, [C]: G. Chen et al. 2018.](/images/stories/EN125/goeltz3.png)
Figure 3: Comparison with other spike-based neuromorphic classifiers on the MNIST data set, see [1] for details. [A]: E. Stromatias et al. 2015, [B]: S. Esser et al. 2015, [C]: G. Chen et al. 2018.
The underlying learning algorithm is built on a rigorous derivation of the spike time in biologically inspired neuronal systems. This makes it possible to precisely quantify the effect of input spike times and connection strengths on later spikes, which in turn allows this effect to be computed throughout networks of multiple layers (Figure 1). The precise value of a single spike’s effect is used on a host computer to calculate a change in the connectivity of neurons on the chip that improves the network’s output. Crucially, we demonstrated that our approach is stable with regards to various forms of noise and deviations from the ideal model, which represents an essential prerequisite for physical computation, be it biological or artificial. This makes our algorithm suitable for implementation on a wide range of neuromorphic platforms.
Although these results are already highly competitive compared to other related neuromorphic realisations of spike-based classification (Figure 3), it is important to emphasise that the BrainScaleS-2 neuromorphic chip is not specifically optimised for our form of neural computation and learning but is rather a multi-purpose research device. It is likely that optimisation of the system, or hardware dedicated to classification alone will further exploit the algorithm’s benefits. Even though the current BrainScaleS-2 generation is limited in size, the algorithm can scale up to larger systems. In particular, coupled with the intrinsically parallel nature of the accelerated hardware, a scaled-up version of our model would not require longer execution time, thus conserving its advantages when applied to larger, more complex data. We thus view our results as a successful proof-of-concept implementation, highlighting the advantages of sparse, but robust coding combined with fast, low-power silicon substrates, with intriguing potential for edge computing and neuroprosthetics.
We would like to thank our collaborators Andreas Baumbach, Sebastian Billaudelle, Oliver Breitwieser, Benjamin Cramer, Dominik Dold, Akos F. Kungl, Walter Senn, Johannes Schemmel and Karlheinz Meier, as well as the Manfred Stärk foundation for ongoing support. This research has received funding from the European Union’s Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 945539 (Human Brain Project SGA3).
Links:
[L1] https://www.youtube.com/watch?v=ygLTW_6g0Bg
[L2] https://physio.unibe.ch/~petrovici/group/
[L3] https://www.kip.uni-heidelberg.de/vision/
References:
[1] J. Göltz, L. Kriener, et al.: “Fast and deep: energy-efficient neuromorphic learning with first-spike times,” arXiv:1912.11443, 2019.
[2] S. Billaudelle, et al.: “Versatile emulation of spiking neural networks on an accelerated neuromorphic substrate,” 2020 IEEE International Symposium on Circuits and Systems (ISCAS). IEEE, 2020.
[3] L. Kriener, et al.: “The Yin-Yang dataset,” arXiv:2102.08211, 2021.
Please contact:
Julian Göltz, NeuroTMA group, Department of Physiology, University of Bern, Switzerland and Kirchhoff-Institute for Physics, Heidelberg University, Germany











