by Andreas Baumbach (Heidelberg University, University of Bern), Sebastian Billaudelle (Heidelberg University), Virginie Sabado (University of Bern) and Mihai A. Petrovici (University of Bern, Heidelberg University)
Uncovering the mechanics of neural computation in living organisms is increasingly shaping the development of brain-inspired algorithms and silicon circuits in the domain of artificial information processing. Researchers from the European Human Brain project employ the BrainScaleS spike-based neuromorphic platform to implement a variety of brain-inspired computational paradigms, from insect navigation to probabilistic generative models, demonstrating an unprecedented degree of versatility in mixed-signal neuromorphic substrates.
Unlike their machine learning counterparts, biological neurons interact primarily via electrical action potentials, known as “spikes”. The second generation of the BrainScaleS neuromorphic system [1, L6] implements up to 512 such spiking neurons, which can be near-arbitrarily connected. In contrast to classical simulations, where the simulation time increases for larger systems, this form of in-silico implementation replicates the physics of neurons rather than numerically solving the associated equations, enabling what could be described as perfect scaling, with the duration of an emulation being essentially independent of the size of the model. Moreover, the electronic circuits are configured to be approximately 1000 times faster than their biological counterparts. It is this acceleration that makes BrainScaleS-2 a powerful platform for biological research and potential applications. Importantly, the chip was also designed for energy efficiency, with a total nominal power consumption of 1 W. Coupled with its emulation speed, this means energy consumption is several orders of magnitude lower than state-of-the art conventional simulations. These features, including the on-chip learning capabilities of the second generation of the BrainScaleS architecture, have the potential to enable the widespread use of spiking neurons beyond the realm of neuroscience and into artificial intelligence (AI) applications. Here we showcase the system’s capabilities with a collection of five highly diverse emulation scenarios, from a small model of insect navigation, including the sensory apparatus and the environment, to fully fledged discriminatory and generative models of higher brain functions [1].
As a first use case, we present a model of the bee’s brain that reproduces the ability of bees to return to their nest’s location after exploring their environment for food (Figure 1). The on-chip general-purpose processor of BrainScaleS is used to simulate the environment and to stimulate the motor neurons triggering the exploration of the environment. The network activity stores the insect’s position, enabling autonomous navigation back to its nest. The on-chip environment simulation avoids delays typically introduced by the use of coprocessors, thus fully exploiting the speed of the accelerated platform for the experiment. In total, the emulated insect performs about 5.5 biological hours’ worth of exploration in 20 seconds.
While accelerated model execution is certainly an attractive feature, it is often the learning process that is prohibitively time-consuming. Using its on-chip general-purpose processor, the BrainScaleS system offers the option of fully embedded learning, thus maximising the benefit of its accelerated neuro-synaptic dynamics. Using the dedicated circuitry for spike-timing-dependent plasticity (STDP), the system can, for example, implement a neuromorphic agent playing a version of the game Pong. The general-purpose processor again simulates the game dynamics and, depending on the game outcome, provides the success signal used in the reinforcement learning. Together with the analogue STDP measurements, this reward signal allows the emulated agent to learn autonomously. The resulting system is an order of magnitude faster and three orders of magnitude more energy-efficient than an equivalent software simulation (Figure 1).
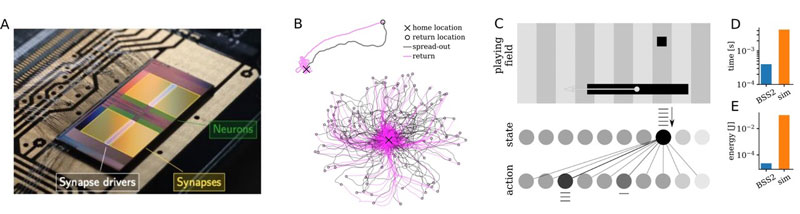
Figure 1: (A) Photograph of the BrainScaleS chip with false-colour overlay of some of its components. (B) 100 paths travelled by an emulated insect swarming out from its nest, exploring the world (grey) and returning home (pink). (C) Emulated Pong agent and associated spiking network. Emulation time and energy consumption compared to state-of-the art simulations shown in (D) and (E).
In a different example of embedded learning, a network is trained to discriminate between different species of iris flowers based on the shape of their petals using a series of receptors in a two-dimensional dataspace (Figure 2). For each neuron, the number of possible inputs is limited by the architecture and topology of the silicon substrate, much like the constraints imposed by biological tissue. Here too, the learning rule is based on spike-time similarities but enhanced by regularisation and pruning of weaker synaptic connections. On BrainScaleS, it is possible to route the output spikes of multiple neurons to each synaptic circuit. This allows the adaptation of the network connectome at runtime. In particular, synapses below a threshold strength are periodically removed and alternatives are instantiated. The label neurons on BrainScaleS then develop distinct receptive fields using the same circuitry. For this particular problem, it was possible to enforce a sparsity of nearly 90% without impacting the final classification performance (Figure 2).
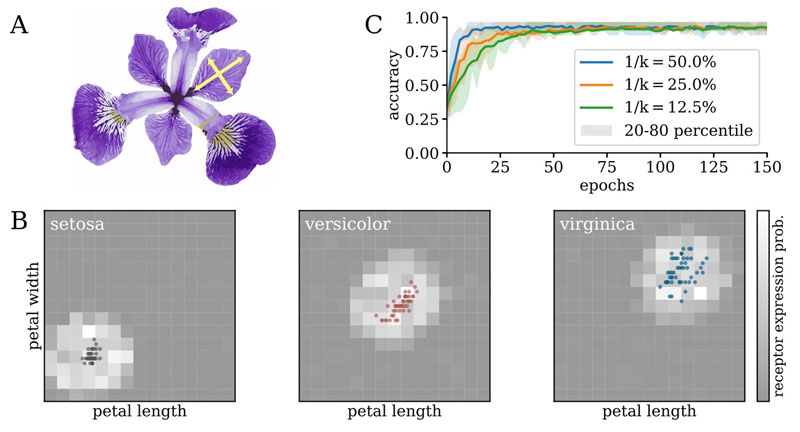
Figure 2: (A) Different species of Iris can be distinguished by the shape of their flowers’ petals. (B) Structural changes to the connectome of an emulated classifier organize appropriate receptive fields even for very sparse connectivity throughout the experiment. (C) Evolution of classification accuracy during training for different sparsity levels.
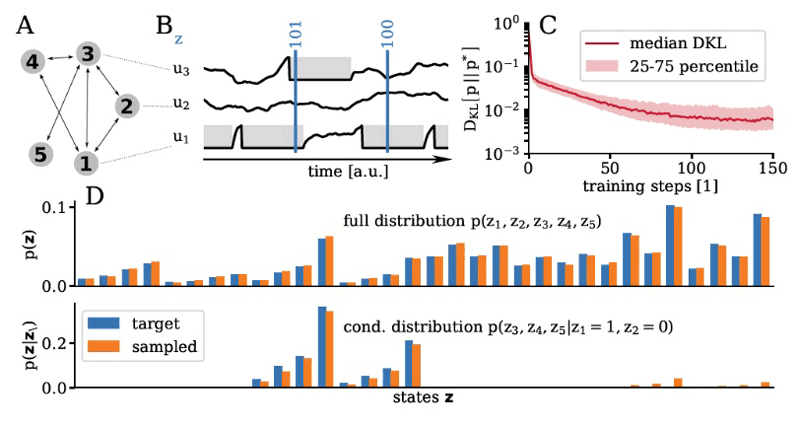
Figure 3: Bayesian inference with spiking neural networks. (A, B) Neuron as spike patterns can be interpreted as binary vectors. (C) Neural dynamics can learn to sample from arbitrary probability distributions over binary spaces (D).
The last two networks demonstrated in [1] hint at the more challenging nature of tasks that biological agents must solve in order to survive. One such task is being able to cope with incomplete or inaccurate sensory input. The Bayesian brain hypothesis posits that cortical activity instantiates probabilistic inference at multiple levels, thus providing a normative framework for how mammalian brains can perform this feat. As an exemplary implementation thereof, we showcase Bayesian inference in spiking networks on BrainScaleS (Figure 3). Similar systems have previously been used to generate and discriminate between hand-written digits or small-scale images of fashion articles [3, 4].
The final network model was trained as a classifier for visual data using time-to-first-spike coding. This specific representation of information enabled it to classify up to 10,000 images per second using only 27 µJ per image. This particular network is described in more detail in its own dedicated article [L5].
These five networks embody a diverse set of computational paradigms and demonstrate the capabilities of the BrainScaleS architecture. Its dedicated spiking circuitry coupled with its versatile on-chip processing unit allows the emulation of a large variety of models in a particularly tight power and time envelope. As work continues on user-friendly access to its many features, BrainScaleS aims to support a wide range of users and use cases, from both the neuroscientific research community and the domain of industrial applications.
We would like to thank our collaborators Yannik Stradmann, Korbinian Schreiber, Benjamin Cramer, Dominik Dold, Julian Göltz, Akos F. Kungl, Timo C. Wunderlich, Andreas Hartel, Eric Müller, Oliver Breitwieser, Christian Mauch, Mitja Kleider, Andreas Grübl, David Stöckel, Christian Pehle, Arthur Heimbrecht, Philipp Spilger, Gerd Kiene, Vitali Karasenko, Walter Senn, Johannes Schemmel and Karlheinz Meier, as well as the Manfred Stärk foundation for ongoing support. This research has received funding from the European Union’s Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 945539 (Human Brain Project SGA3).
Links:
[L1] Presentation to [1]: https://youtu.be/_x3wqLFS278
[L2] NeuroTMA group: https://physio.unibe.ch/~petrovici/group/
[L3] Electronic Visions group: https://www.kip.uni-heidelberg.de/vision/
[L4] HBP neuromorphic computing platform: https://electronicvisions.github.io/hbp-sp9-guidebook/
[L5] ERCIM News article: “Fast and energy-efficient deep neuromorphic learning”
[L6] ERCIM news article: “The BrainScaleS Accelerated Analogue Neuromorphic Architecture”
References:
[1] S. Billaudelle, et al.: “Versatile emulation of spiking neural networks on an accelerated neuromorphic substrate.” 2020 IEEE International Symposium on Circuits and Systems (ISCAS). IEEE, 2020.
[2] T. Wunderlich, et al.: “Demonstrating advantages of neuromorphic computation: a pilot study,” Frontiers in Neuroscience,vol. 13, p. 260, 2019.
[3] D. Dold, et al. “Stochasticity from function—why the bayesian brain may need no noise.” Neural networks 119 (2019): 200-213.
[4] A. Kungl, F. Akos, et al.: “Accelerated physical emulation of Bayesian inference in spiking neural networks”, Frontiers in neuroscience 13 (2019): 1201.
Please contact:
Andreas Baumbach, NeuroTMA group, Department of Physiology, University of Bern, Switzerland and Kirchhoff-Institute for Physics, Heidelberg University, Germany











