by Michaela Areti Zervou, Effrosyni Doutsi, Panagiotis Tsakalides (University of Crete and FORTH-ICS)
Precision medicine holds the promise of personalised treatment based on an individual's genetic makeup. However, the lengthy and costly process of drug discovery hinders progress. Can artificial intelligence (AI), specifically generative models offer a solution? Is it possible to efficiently select the most promising drug candidates for validation? Our research focuses on developing robust tools that streamline the validation process of drug design – saving time and resources.
Precision medicine [1] represents a new era in health care, aiming to provide tailored treatments based on an individual's unique genetic composition. By analysing an individual's DNA, healthcare providers can make decisions about the most suitable treatment. However, significant challenges in developing new drugs, such as the time consumption and the considerable cost of the design process, often involves years of trial and error with millions of compounds being tested.
A crucial task for efficient drug design is accurately predicting the position of the target protein within the cell. This knowledge empowers the design of drugs that specifically target an intended protein, disrupt or enhance its function while minimising the risk of unintended effects on healthy cells. This holds the key to developing highly targeted drug candidates that can bind with exceptional precision, maximising therapeutic benefits, while reducing potential adverse reactions.
The power of artificial intelligence (AI) offers promising opportunities to address the challenges in predicting protein position and designing drugs with enhanced specificity. But how can we leverage the power of AI to predict protein position and design drugs that specifically target intended proteins while minimising off-target effects? The answer lies in the realm of generative models [2]. These models are a fascinating blend of mathematics, statistics and computer engineering. They are trained on large databases of existing molecules trying to identify and learn the underlying patterns and relationships between molecular structures. Based on the acquired knowledge, they allow generation of a vast number of novel molecules that adhere to the learned patterns while introducing variations. This enables researchers to explore a wide range of potential drug candidates, but how can we ensure the reliability and effectiveness of all these generated molecules? Moreover, is it possible to accelerate the validation process in the laboratory?
To address these challenges, our research funded by the Hellenic Foundation for Research and Innovation (HFRI) under the Ph.D. Fellowship grant no. 5647 [L1], focuses on developing robust classification tools to effectively learn and classify the position of the proteins in the cell. As shown in Figure 1, by integrating these tools into the de novo design pipeline, we can guide the process of designing to produce novel molecules with a desired position of the target protein in the cell. This approach enables researchers to prioritise and select the most promising candidates for experimental validation, thereby streamlining the validation process. Ultimately, this reduces the number of potential designs that require synthesis and testing, leading to significant time and resource savings.
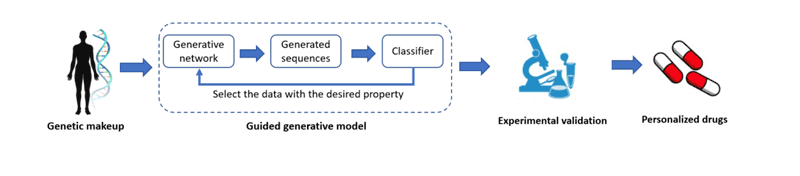
Figure 1: Personalised drug-design architecture pipeline.
Our research team has made significant advancements in the development of novel classification tools that surpass the current state-of-the-art [3]. Leveraging innovative algorithms and machine learning techniques, we have created highly accurate tools that can effectively identify the position of the target protein in the cell.
Furthermore, to bridge the gap between research and practical applications, we have established collaborations with renowned biologists and experts in the field. These collaborations enable us to validate the effectiveness of our classification tools in real-world scenarios, leveraging their domain knowledge and expertise. By aligning our research with industry requirements and working closely with experienced biologists, we aim to develop practical solutions that have the potential to revolutionise the drug-discovery process.
In conclusion, precision medicine holds immense potential for personalised health care. However, the lengthy and costly drug-discovery process poses significant challenges. The power of artificial intelligence and generative models presents an opportunity for a breakthrough in this domain. Efficiently selecting the most promising drug candidates for validation is crucial to saving time and resources. Through our research and collaborations with experts in the field, including renowned biologists, we are working towards a more efficient and effective drug-design process. By harnessing the capabilities of artificial intelligence and leveraging generative models, we aim to revolutionise the development of personalised treatments. Ultimately, our efforts bring us one step closer to realising the transformative impact of personalised medicine on patients worldwide.
Links:
[L1] https://www.elidek.gr/en/call/3rd-call-for-h-f-r-i-scholarships-for-phd-candidates/
References:
[1] M.R. Kosorok and E. B. Laber, “Precision medicine,” Annual Review of Statistics and its Application, vol. 6, pp. 263–286, 2019.
[2] X. Pan and T. Kortemme, “Recent advances in de novo protein design: principles, methods, and applications,” Journal of Biological Chemistry, vol. 296, 2021.
[3] M. A. Zervou, E. Doutsi and P. Tsakalides, “Efficient protein structural class prediction via chaos game representation and recurrent neural networks,” 2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Rhodes Island, Greece, 2023, pp. 1–5, doi: 10.1109/ICASSP49357.2023.10094877.
Please contact:
Michaela Areti Zervou , University of Crete and FORTH-ICS, Greece
Effrosyni Doutsi , FORTH-ICS, Greece











