by Markus Hittmeir, Rudolf Mayer and Andreas Ekelhart (SBA Research)
The microbial communities on the human body are subject to extensive research. While individual variations in the microbiome reveal valuable information about health and diseases, they also allow for the identification of individuals among populations of hundreds. The resulting demand for solutions to protect the privacy of participants in microbiome studies can be met by adapting well-known anonymisation techniques.
The bacteria, fungi and protists living on various sites of the human body have a substantial influence on our wellbeing. Studies of the human microbiome can help us with the prediction, diagnosis and treatment of diseases, and new findings are published on a regular basis. For instance, changes in the gut microbiome may be related to gastrointestinal diseases, obesity, diabetes, and depression [1]. As more data on the microbiome is gathered and stored, investigations into the temporal and individual stability of microbiome readings and the ensuing privacy risks have gained importance.
In 2015, Franzosa et al. presented a method for the unique characterization of hundreds of individuals via short codes constructed from their microbiome samples [2]. Using follow-up samples collected between 30 and 300 days later, about 30% of the individuals could still be matched correctly by comparing the samples’ codes. While this result is the average of several body sites, the gastrointestinal microbiome appeared to be exceptionally stable and allowed the researchers to match up to 80% of individuals. The authors concluded that their work demonstrates the feasibility of microbiome-based identifiability, which poses ethical implications for the design of microbiome studies and a need for privacy-enhancing solutions for microbiome data. Recently, this demand has been strengthened by an improvement of Franzosa et al.’s technique [3], leading to an increased number of individuals that can be re-identified based on their microbiome.
In order to give an overview of the new method in [3] and its differences to [2], let us start by taking a closer look at the microbiome data. In addition to the aforementioned gastrointestinal microbiome, samples may be taken from several other body sites, such as saliva, throat, anterior nares (the external portion of the nose), supragingival plaque (at the teeth) or buccal mucosa (at the inside of the cheek). Starting with large volumes of raw data containing the genetic sequences of microbes found in the sample, there are several possibilities for the subsequent feature extraction. One method is to measure the abundance of bacterial and archaeal species found in the sample, leading to a table similar to the excerpt shown in Figure 1. The rows refer to the various species, and the columns (the “sample vectors”) contain the abundance counts for the individual samples. The relative counts in Figure 1 are proportions, meaning that the sum of all values in each column equals 1. Full examples for such datasets can be found under [L1], together with an implementation of the method in [2].
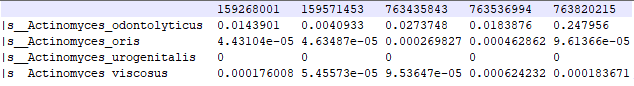
Figure 1: Excerpt from a microbiome table with features based on relative species abundance. The nine-digit number in the first row is the identifier of the individuals of the study.
While there are publicly available techniques [L2] for microbiome-based identification on the raw genetic data, both [2] and [3] focus on privacy risks that arise from datasets containing sample vectors as discussed above. For each such sample, Franzosa et al. consider the features as either present or absent, based on a threshold (e.g., 0.0001) for the abundance. The code of each sample is then a unique combination of its present features, and the experiments in [2] demonstrate their temporal stability. The improvement in [3] is based on considering not just a subset of the present features, but comparing complete sample vectors. In order to match a single sample against a whole dataset, the method computes its distance to all the columns and finds the closest one (the “nearest-neighbour”). Compared to [2], this leads to an improved identification on most of the considered datasets. In particular, we see an increase in the average percentage of true-positive matches of 28% on the widely studied gut microbiome. In addition, the introduction of a criterion for accepting neighbouring pairs of samples as possible matches prevents a large number of false positives (i.e., incorrect matches). Figure 2 shows the results on six different body sites.
![Figure 2: Re-identification results in % of the approach in [3] on six different body sites. On each body site, the left bar displays the results without acceptance criterion. After applying the criterion, we can see that most false positives turn into false negatives, improving the accuracy of the technique. Figure 2: Re-identification results in % of the approach in [3] on six different body sites. On each body site, the left bar displays the results without acceptance criterion. After applying the criterion, we can see that most false positives turn into false negatives, improving the accuracy of the technique.](/images/stories/EN126/hitmaier2.png)
Figure 2: Re-identification results in % of the approach in [3] on six different body sites. On each body site, the left bar displays the results without acceptance criterion. After applying the criterion, we can see that most false positives turn into false negatives, improving the accuracy of the technique.
The threat analysis conducted in [2] and [3] demonstrates that the extent of the privacy risk depends on factors such as feature types and body sites. In this context, an adversary is any party in possession of unidentified microbiome samples with the intention to link them to other samples for accumulating information about the underlying individual, such as the participation in a specific study, or metadata linked to the identified record. There are multiple avenues by which an adversary could obtain microbiome samples, including public databases, cyberattacks against healthcare facilities and research organisations, data exfiltration via insiders, and potentially, directly from the victim (e.g., saliva).
One solution for protecting a microbiome database D is to establish k-anonymity, meaning that groups of at least k samples in D are indistinguishable to the discussed identification techniques. It is then impossible to find unique matches, and an adversary has to guess the correct individual from at least k different choices. This goal may be achieved by adapting a variety of classical techniques for k-anonymity on relational data. Let us briefly consider one such idea for establishing 2-anonymity. One first computes the pairwise distances between all the samples in D and finds pairs of samples that are most similar. Next, each pair is generalised by computing the mean of all the abundance counts of the two samples. Finally, each original sample in D is replaced by the generalisation of its corresponding pair, leading to 2-anonymity. Note that k-anonymity may be achieved by considering clusters instead of pairs. Moreover, there are several possibilities to optimise the procedure and minimise the information loss. In this sense, future work will focus on the refinement of techniques for mitigating the capabilities of an adversary and, thus, the related risks.
This work was partially funded by the Austrian Research Promotion Agency FFG under grant 877173 (GASTRIC).
Links:
[L1] https://huttenhower.sph.harvard.edu/idability
[L2] https://github.com/princello/GePMI
References:
[1] G. Rogers, et al.: “From gut dysbiosis to altered brain function and mental illness: mechanism and pathways”, Mol Psychiatry 21, 738-748, 2016.
[2] E. Franzosa, et al.: “Identifying personal microbiomes using metagenomic codes”, PNAS 112, E2930-E2938, 2015.
[3] M. Hittmeir, et al.: Distance-based techniques for personal microbiome identification, 2021, under review.
Please contact:
Markus Hittmeir, Rudolf Mayer and Andreas Ekelhart
SBA Research gGmbH, Austria
{mhittmeir, rmayer, aekelhart}@sba-research.org











