by Bart Kamphorst (TNO), Daan Knoors (IKNL) and Thomas Rooijakkers (TNO)
Researchers in oncology require comprehensive patient data to reflect on cancer care and prevention. However, given the complexity of cancer, some research questions require patient data that is distributed over multiple registries, and it can be challenging to access or exchange such highly sensitive health data. To get around this problem, the Netherlands Comprehensive Cancer Organisation (IKNL) and the Netherlands Organisation for Applied Scientific Research (TNO) have collaboratively developed algorithms that enable survival analyses on distributed data with rigorous privacy guarantees.
The Netherlands Comprehensive Cancer Organisation (IKNL) maintains the Netherlands Cancer Registry (NCR) to enable healthcare professionals, researchers and policymakers to reflect on cancer care and prevention. Even though the NCR is one of the largest disease-specific registries in the world, given the complexity of cancer, some research questions require additional information that is collected by other organisations. For instance, studies that aim to understand which factors influence a patient's chance of survival could reveal new patterns when additional information is considered, like drug usage, hereditary conditions, or comorbidities.
Clinical studies benefit from having as much relevant data as possible. However, organisations are prevented from combining data due to privacy concerns, regulations, and other factors. Particularly, sensitive patient-level data must always be treated with great care. On the other hand, population-level insights that can be obtained from analysing patient-level data might be less sensitive. As a result, over recent decades, new methods have been investigated that obtain population-level insights while providing strong guarantees that the private information of patients is protected. IKNL and the Netherlands Organisation for Applied Scientific Research (TNO) have designed and implemented such privacy-preserving methods in the domain of survival analysis.
Survival analysis – analysing the expected time it takes for an event, such as death, hospitalisation, or tumour recurrence, to occur – is an important aspect of oncological research. Survival analysis can be used to indicate the likelihood of someone being alive a few years after diagnosis. Additionally, it can give insights into which characteristics might relate to the chances of survival, e.g., the patient’s fitness, the treatment method, and hospital of diagnosis.
An often-used survival analysis technique is the Kaplan–Meier estimator, a non-parametric statistic used to estimate the survival function of a lifetime table. To compare survival between groups, we can use the log-rank test associated with the Kaplan-Meier estimator. The log-rank test is a statistical procedure that compares two or more survival distributions. A direct application is to test whether one treatment has a greater effect on the longevity of a patient compared to another. Advances in machine learning and cryptography allow us to compute this log-rank statistic without disclosing any underlying patient-level information.
IKNL is leading the development of an infrastructure called vantage6 [L1], which enables organisations to jointly perform analyses without needing to share their respective data. This federated-learning based approach works well if the data is horizontally distributed, e.g., the participating organisations maintain the same type of data of different patients. In the Kaplan-Meier setting, this translates to organisations that all record the patient group, the outcome of the experiment and the time of that outcome for separate patients. Vantage6 facilitates privacy-preserving analyses on the combined sets of patients, leveraging the fact that every organisation can perform the analysis on the data of its own patients.
An alternative scenario is that data is vertically distributed, e.g., participating organisations maintain different, complementary types of information about the same patient. In our setting this translates to one organisation recording the patient group (e.g., based on comorbidities), and another organisation recording the survival data. Computing the log-rank statistic, however, requires knowledge of both the patient group information and the survival data. Lacking either type of data makes it impossible to deduce any meaningful insights. Using the cryptographic concepts of Secure Multi-Party Computation (MPC), it is possible to perform the analysis in this scenario while preserving the patients’ privacy.
MPC is a set of techniques that enables multiple entities to jointly evaluate a function on their data, without revealing that data to one another. Some techniques achieve this property by supporting computations on encrypted data (e.g., homomorphic encryption), which particularly enables computations that involve the sensitive but encrypted data of another entity, whereas some other techniques (e.g., secret sharing) split the sensitive data in multiple pieces in such a way that computations can be performed on the separate pieces. Most importantly, within some specified security model, every MPC technique guarantees to preserve privacy throughout the entire computation.
In our joint 2020 research project, we have developed and implemented new MPC solutions to compute the log-rank statistic of the Kaplan-Meier estimator on vertically-distributed data. These privacy-preserving solutions do not reveal the group information and the survival data to anyone. Experiments show that the solutions are sufficiently fast and scalable to be used in real-world settings. The protocol is visualised in Figure 1. An open-source implementation is provided on GitHub [L2]. Our protocol does not reveal the Kaplan-Meier estimators themselves since patient-level information can be deduced from them. Presenting the Kaplan-Meier estimators in a more privacy-friendly way is described in Vogelsang et al. [1].
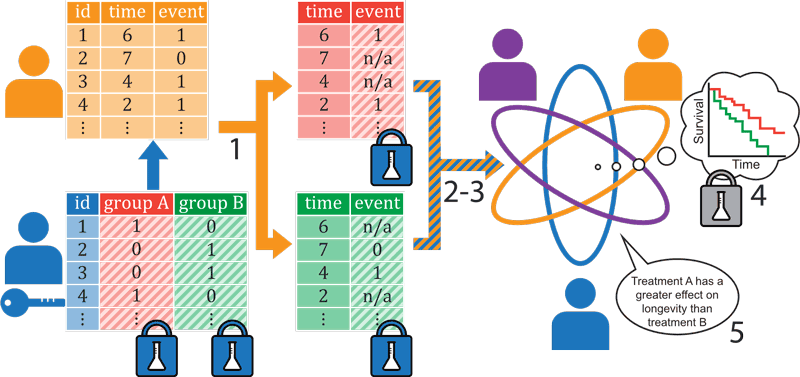
Figure 1: The protocol to securely compute the log-rank statistic for vertically-partitioned data. One party (Blue) owns data on patient groups, the other party (Orange) owns data on event times (did the patient experience an event ‘1’ or not ‘0’, and when did this occur). Protocol outline: Blue encrypts its data using additive homomorphic encryption and the encrypted data is sent to Orange. Orange can securely, without decryption, split its data in the patient groups specified by Blue (1) using the additive homomorphic properties of the encryptions. Orange performs some preparatory, local, computations (2) and with the help of Blue secret-shares the data (3) between Blue, Orange and Purple, where Purple is introduced for efficiency purposes. All parties together securely compute the log-rank statistic associated with the (never revealed) Kaplan-Meier curves (4) and only reveal the final statistical result (5).
Motivated by these promising results, TNO and IKNL developed other relevant algorithms to enable privacy-preserving survival analyses, including Cox Proportional Hazard. Future activities include the extension of the toolkit to other relevant privacy-preserving machine learning algorithms for medical analyses in the cancer domain.
Links:
[L1] https://vantage6.ai/
[L2] https://github.com/TNO-MPC/protocols.kaplan_meier
References:
[1] Vogelsang et al.: “A Secure Multi-Party Computation Protocol for Time-To-Event Analyses", in Studies in health technology and informatics, 270, 2020,
doi: 10.3233/SHTI200112.
Please contact:
Bart Kamphorst
TNO, the Netherlands











