by Massimiliano Assante (CNR-ISTI), Marco Lettere (Nubisware srl), Alfredo Oliviero (CNR-ISTI), and Pasquale Pagano (CNR-ISTI)
The D4Science platform is advancing reproducible research by providing scientists with robust, cloud-based tools for large-scale data analysis such as the Cloud Computing Platform (CCP). CCP enhances collaboration, allowing researchers to share, reuse, and build on each other’s work across diverse scientific disciplines.
D4Science [1, 2] embraces the “as a Service” paradigm, offering Virtual Research Environments (VREs) [3] to streamline the research process, serving as a foundation for modern scientific collaboration, combining accessibility, innovation, and scalability within a single, cohesive framework. The VREs allow researchers to perform their data-driven research tasks without needing to manage the complexities of storage, computation, or deployment.
The Cloud Computing Platform (CCP) [L1], born from D4Science’s more than 10 years of experience as an operational digital infrastructure, embodies the principles of FAIR (Findable, Accessible, Interoperable, and Reusable) data, advancing Open Science and reproducibility in research. At its core, CCP is designed to handle large-scale data analysis, promoting the widespread adoption of microservice-based architectures. This approach enhances the platform’s flexibility and makes it highly interoperable and composable, enabling researchers to build and integrate their computational methods effortlessly. CCP incorporates several innovative features that make it particularly suited for data-intensive research. Its methods importer tool simplifies the integration of computational methods, allowing users to deploy custom algorithms and applications. The execution lifecycle tracker ensures that every step of a method’s life, from creation to execution, is meticulously documented. Additionally, CCP includes a real-time execution monitor, which provides live feedback from the server logs during method execution, enabling users to track progress and identify issues promptly. Furthermore, CCP supports the archiving of executions, preserving the full configuration, parameters, and inputs of each execution. This capability ensures that methods can be repeated with fidelity in the future, enabling reproducibility and providing a robust framework for iterative research processes.
One of CCP’s core strengths lies in its flexibility, enabling seamless integration across programming environments, languages, and execution infrastructures. This flexibility extends to the execution of methods by the adoption of standard REST/JSON APIs for interacting with CCP. Methods can be executed programmatically from web applications as shown in Figure 1, command-line interfaces, Jupyter Notebooks or Galaxy workflows. To further support users, automatic code generators create stubs and templates for multiple languages and runtimes, including Python, Julia, Bash, Galaxy, and Notebooks.
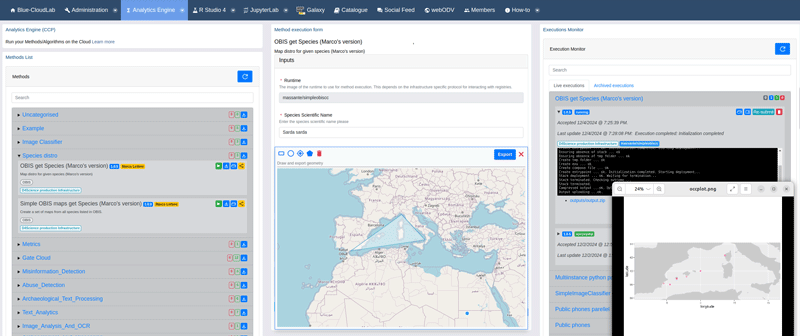
Figure 1 - The Cloud Computing Platform Web User Interface
CCP supports both containerised (e.g., Docker, Docker Swarm, LXD, Kubernetes, Singularity) and non-containerised infrastructures (e.g., Galaxy, Slurm). Execution infrastructures can be hosted on commercial platforms such as Google Cloud Platform, or non-commercial ones like D4Science’s production environment. They also accommodate High-Performance Computing (HPC) clusters, as well as local environments such as personal laptops for experimental purposes.
The flexible and inherently distributed nature of CCP execution infrastructures allows users to design and run methods precisely aligning computational workloads with the most suitable resources thus fostering optimisation of execution environments based on data locality, computational demand and infrastructure capabilities. By accommodating a broad spectrum of execution contexts, from HPC systems to experimental environments, CCP facilitates distributed and scalable collaboration, enabling reproducibility and interoperability across diverse research domains.
D4Science offers a native execution infrastructure based on Docker swarm and enriched by a dedicated image registry based on Harbor. Especially when designing methods for such container-based infrastructures, scientists have virtually no limitations on what programming languages, environments, versions, or dependencies they are allowed to use. By encapsulating computational methods and their dependencies into isolated containers, CCP ensures reproducibility, portability, and scalability. This approach allows researchers to deploy methods without compatibility concerns, maintaining consistent execution environments regardless of underlying hardware or software variations. Additionally, the use of containers significantly reduces the overhead associated with traditional virtualisation technologies, thereby improving the efficiency of complex scientific workflows.
By integrating containerisation, flexible infrastructure management, and robust automation tools, CCP emerges as a critical enabler of large-scale distributed computing, driving innovation and reproducibility across research disciplines.
In addition to its technical features, CCP strongly aligns with the principles of Open Science, ensuring that all scientific outputs are transparent, repeatable, and reusable. Every execution within CCP is documented with comprehensive provenance tracking, which records the origins, transformations, and outcomes of data. This capability not only supports reproducibility but also provides a clear lineage for scientific discoveries, uplifting trust and attribution in research. This flexibility ensures that researchers across disciplines, regardless of their preferred tools, can adopt and benefit from the platform. Furthermore, the integration of dynamic resource allocation allows users to scale their computational resources based on demand, making CCP suitable for projects of varying sizes and complexities.
The platform’s impact is already visible in large-scale scientific initiatives. For instance, the EOSC Blue-Cloud2026 project [L2] leverages CCP for ocean science research, utilising its robust infrastructure to perform collaborative data analytics on vast datasets. Similarly, the SoBigData Research Infrastructure [L3], focusing on social data mining and ethical Big Data analytics, integrates CCP to enable a multidisciplinary ecosystem for studying social phenomena. These examples highlight how CCP empowers researchers to address complex scientific questions by providing the tools necessary for effective collaboration, computation, and discovery.
By bridging the gap between technical complexity and scientific innovation, with its combination of containerisation, API-driven workflows, provenance management, and scalable infrastructure, CCP offers researchers a reliable and efficient environment for advancing their work. As scientific challenges grow in complexity, platforms like CCP will play an increasingly critical role in enabling reproducible and impactful research across disciplines.
Links:
[L1] https://ccp.cloud.d4science.org/docs/index.html
[L2] https://www.blue-cloud.org
[L3] http://www.sobigdata.eu
References:
[1] M. Assante, et al, “Enacting open science by D4Science,” Future Gener. Comput. Syst., vol. 101, pp. 555–563, 2024, doi: 10.1016/j.future.2019.05.063.
[2] L. Candela, D. Castelli, and P. Pagano, “The D4Science Experience on Virtual Research Environment Development,” Comput. Sci. Eng., vol. 25, no. 2, pp. 12–19, 2023, doi: 10.1109/MCSE.2023.3290433.
[3] M. Assante, et al., “Virtual research environments co-creation: The D4Science experience,” Concurr. Comput. Pract. Exp., vol. 35, no. 18, pp. e6925:1–e6925:12, 2023, doi: 10.1002/cpe.6925.
Please contact:
Massimiliano Assante
CNR-ISTI, Italy











