by Olivier Colliot (CNRS), Fabrizio De Vico Fallani and Stanley Durrleman (Inria)
Neurodegenerative diseases, such as Alzheimer’s and Parkinson’s disease, are complex multi-faceted disorders involving a mosaic of alterations that can be measured in the brain of living patients thanks to the tremendous progresses of neuroimaging. A major challenge for computational imaging is to build efficient and meaningful models of disease from multimodal imaging datasets in order to better understand diseases, improve diagnosis and develop precision medicine.
Neurodegenerative diseases, which involve various types of structural and functional alterations in the brain, are a major public health concern. The disease course spans several decades, with a long silent phase in which alterations accumulate without producing symptoms. Finally, neurodegenerative diseases are heterogeneous entities: there is huge variability among patients in terms of pathological alterations, pace of progression and clinical presentation. All these elements hamper the development of new treatments. Computational and statistical approaches have a major role to play to disentangle the complexity of neurodegenerative diseases.
The continuous progress of medical imaging is offering us an increasingly complete picture of brain alterations. Anatomical magnetic resonance imaging (MRI) assesses the integrity of anatomical structures. Diffusion MRI allows the connections between distant brain areas to be mapped in vivo. Functional imaging (functional MRI, electro- and magneto-encephalography) maps neuronal activity. Positron emission tomography (PET) provides information about specific cellular and molecular processes. In recent years, large datasets of patients with multiple imaging modalities and long-term longitudinal follow-up have been gathered. The current challenge is to use these large multimodal datasets to learnmodels of neurodegenerative disease and computer-assisted diagnosis, prognosis and stratification tools.
ARAMIS [L1] is a multidisciplinary research team including computer scientists, specialised in computational imaging and statistical learning, and physicians who specialise in neurodegenerative diseases. It belongs to the Brain and Spine Institute (ICM) [L2], a major research centre on neurological diseases located with the Pitié-Salpêtrière hospital in Paris, the largest adult hospital in Europe, and is jointly affiliated with Inria, CNRS, Inserm (national institute of health) and Sorbonne Universités. ARAMIS aims to build models of neurodegenerative diseases from multimodal brain images. In this paper, we briefly present our most recent results and current projects:
- Comprehensive modelling of brain structure. We recently introduced a comprehensive statistical modelling of the variability of brain structure across patients [1]. This approach integrates grey matter structures, represented as surfaces, and white matter tracks, composed of multiple curves, that connect distant areas. Based on a Bayesian setting, the approach estimates an average model of a population together with its variability (see Figure 1). It can be subsequently used to study the relationship between brain structure alterations and symptoms (as measured by clinical or cognitive tests) or other biological measurements.
- Modelling functional networks. Functional integration of distant brain areas is a central mechanism underlying cognitive tasks which is selectively disrupted by different neurodegenerative diseases. It can be assessed from functional imaging through measurement of temporal dependence between the activity of distant areas. Our team is developing approaches based on the theory of complex networks that can characterise global and local topological properties of functional networks [2]. Such approaches allow network disruptions caused by diseases to be studied and provide new markers for diagnosis and prognosis.
- Learning spatio-temporal models of disease progression. Neurodegenerative diseases evolve during long periods of time, spanning several decades. Modelling the progression of alterations is crucial to predict the evolution of patients and define optimal therapeutic targets. We recently introduced a generic spatio-temporal mixed-effects model which estimates progression trajectories as geodesics on a Riemannian manifold [3]. Importantly, it does not require the definition of arbitrary disease stages but rather estimates time-reparameterisations that account for variability in timing and pace. The model was applied to modelling progression of different types of cognitive and anatomical alterations in Alzheimer’s disease (see Figure 2).
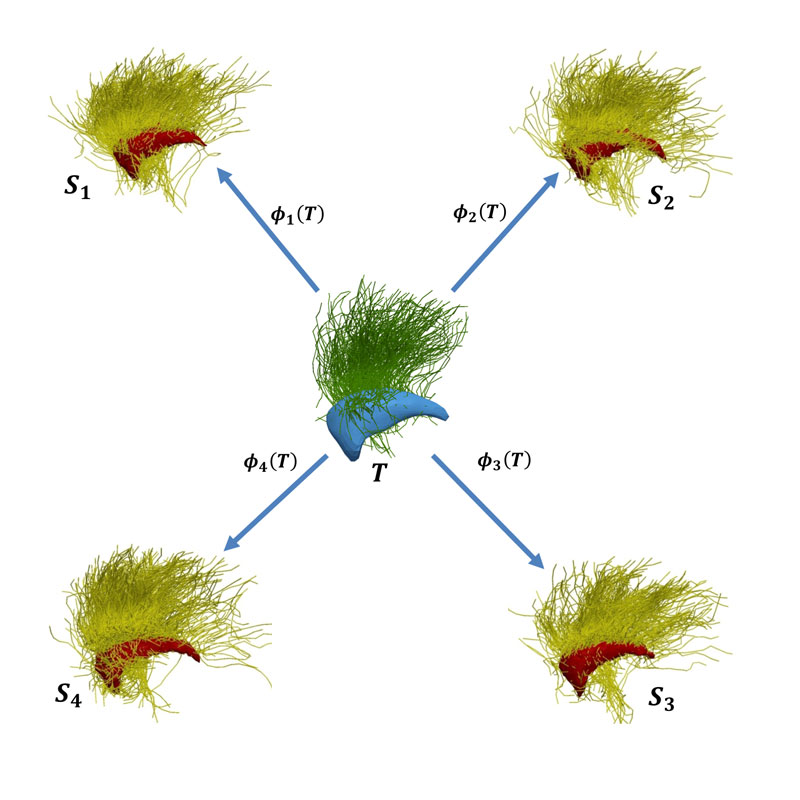
Figure 1: Modelling of brain structure. An average model (called template, denoted as T) is estimated from a population of subjects Si. Each subject is seen as the deformation Φi(T) of the template. The model includes both grey matter structures (surfaces extracted from anatomical MRI) and white matter tracks (curves extracted from diffusion MRI). For the sake of visualisation, only one surface and a set of curves are displayed in the figure.
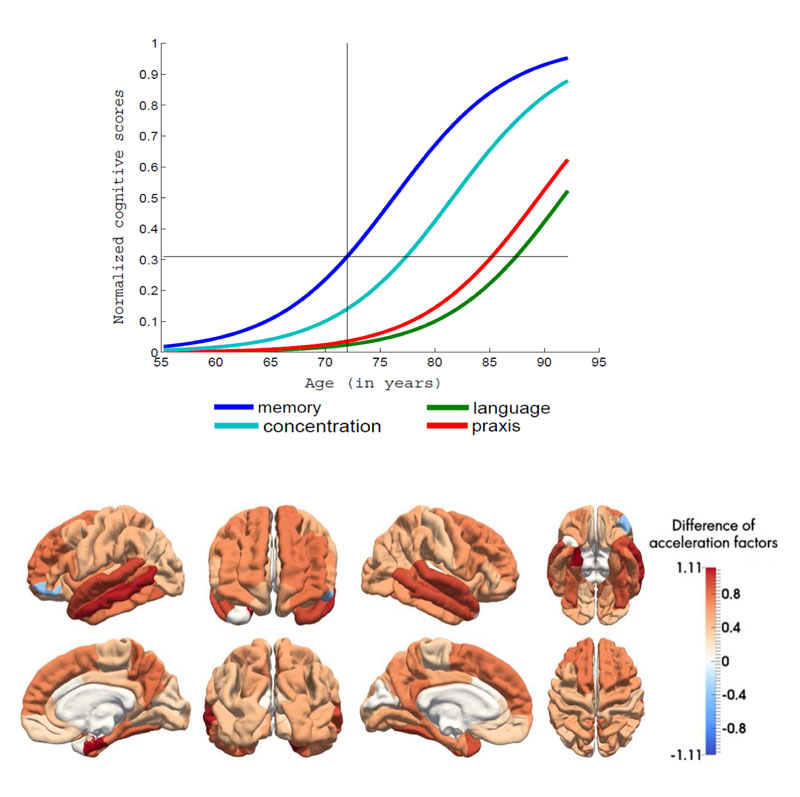
Figure 2: Estimation of disease progression models from a longitudinal dataset of patients with Alzheimer’s disease. Upper panel: average trajectories of data change for a set of cognitive measures. The model allows to infer which types of cognitive deficits occur first. Lower panel: differences of pace of atrophy (extracted from anatomical MRI) between patients with Alzheimer’s disease and healthy controls. This unveils brain regions in which the degenerative process is the more rapid.
Current projects include the definition of network models of temporal progression, the integration of functional imaging (fMRI, PET, EEG/MEG) together with models of brain structure and the design of computer-aided diagnosis and prognosis systems. We are also strongly involved in clinical research studies in different neurodegenerative disorders (Alzheimer’s disease, frontotemporal dementia, primary progressive aphasia) to demonstrate the medical relevance of the developed models. Ultimately, these models should pave the way to digital precision medicine, in which medical decisions can be informed by numerical models personalised to each patient.
Sources of funding
This work is partially funded by European Union’s Horizon 2020 research and innovation programme under grant agreement No 666992, by program “Investissements d’avenir” (ANR-10-IAIHU-06), by NSF-NIH-ANR NetBCI project (ANR-15-NEUC-0006-02) and by the European Research Council (ERC) to SD under grant agreement No 678304.
Links:
[L1] https://www.inria.fr/en/teams/aramis
[L2] http://icm-institute.org/menu/foundation/mission?lang=en
References:
[1] P. Gori et al.: “A Bayesian framework for joint morphometry of surface and curve meshes in multi-object complexes,” Med. Image Anal., vol. 35, pp. 458–474, Jan. 2017.
[2] F. De Vico Fallani, J. Richiardi, M. Chavez, S. Achard: “Graph analysis of functional brain networks: practical issues in translational neuroscience”, Philos. Trans. R. Soc. Lond. B. Biol. Sci., vol. 369, no. 1653, Oct. 2014.
[3] J.-B. Schiratti, S. Allassonniere, O. Colliot, S. Durrleman: “Learning spatiotemporal trajectories from manifold-valued longitudinal data”, in Neural Information Processing Systems, Montréal, Canada, 2015.
Please contact:
Olivier Colliot
CNRS, France
Fabrizio De Vico Fallani,
Stanley Durrleman
Inria, France











