by Erwan Zerhouni, Bogdan Prisacari, Maria Gabrani (IBM Zurich) and Qing Zhong and Peter Wild (Institute of Surgical Pathology, University Hospital Zurich)
Cognitive computing (in the sense of computational image processing and machine learning) helps address two of the challenges of histological image analysis: the high dimensionality of histological images, and the imprecise labelling. We propose an unsupervised method of generating representative image signatures that are robust to tissue heterogeneity. By integrating this mechanism in a broader framework for disease grading, we show significant improvement in terms of grading accuracy compared to alternative supervised feature-extraction methods.
Disease susceptibility and progression is a complex, multifactorial molecular process. Diseases, such as cancer, exhibit cellular heterogeneity, impeding the differentiation between diverse stages or types of cell formations, such as inflammatory response and malignant cell transition. Histological images, that visualise tissues and their cell formations, are huge; three to five orders of magnitude larger than radiology images. The high dimensionality of the images can be addressed via summarising techniques or feature engineering. However, such approaches can limit the performance of subsequent machine learning models to capture the heterogeneous tissue microenvironment. Image analysis techniques of tissue specimens should therefore quantitatively capture the phenotypic properties while preserving the morphology and spatial relationship of the tissue microenvironment.
To capture the diverse features of heterogeneous tissues in large tissue images, we enhance the computational framework introduced by Zerhouni et al [1] by also addressing the cellular heterogeneity, without the need for cellular annotation. This reduces the dependency on labels that tend to be imprecise and tedious to acquire. The proposed method is based on an autoencoder-architecture [2] that we have modified and enhanced to simultaneously produce representative image features as well as perform dictionary learning on these features to reduce dimensionality. We call our method DictiOnary Learning Convolutional autoEncoder (DOLCE).
The principle behind this dimensionality reduction is as follows. DOLCE aims to detect and learn the main morphological and colour patterns of cells that appear in tissue, especially in heterogeneous tissue. The objective is then to represent a cell by combining the main patterns that describe it. By using soft assignments that map each patch of a histology image to a set of dictionary elements, we enable a finer-grained representation of the data. To this end, we can quantify both similarities and differences across patches. In this manner we are able to identify the dominant patterns (deconvolutional filters) that comprise the diverse cell types within a stained tissue type, potentially also across stainings, as well as discriminant patterns, i.e, patterns that are used for very specific patches.
We cross-validate the sparsity of the dictionary representations, in order to find the optimum for each staining. The number of atoms that are needed to robustly represent the sample is staining dependent, thus, indicating a level of heterogeneity. Figure 1 shows a subset of the deconvolutional filters based on hematoxylin and eosin (H&E) staining along with an example tissue sample. As seen from this figure, deconvolutional filters resemble a round shape detector which more likely acts as a cell detector. Even though visualising the learned filters in a network is still an open research problem, one can see that these filters capture local morphology as well as colour information.
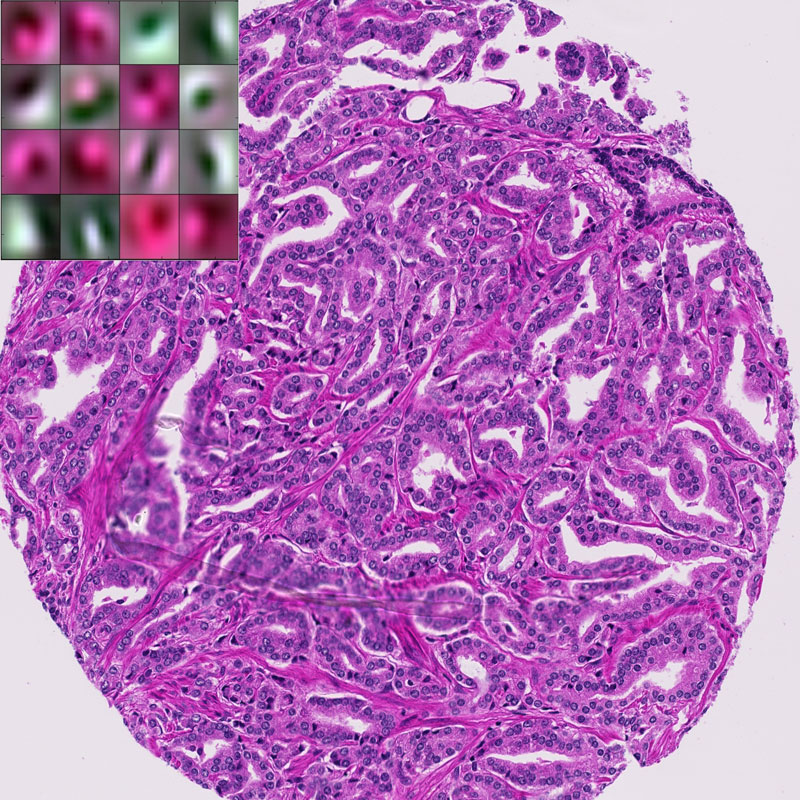
Figure 1: Example of H&E stained prostate tissue with a subset of the learned filters.
To evaluate the proposed framework we tested DOLCE in a prostate cancer dataset and demonstrated its efficacy both in dimensionality reduction and disease grading, as compared to state-of-the-art supervised learning algorithms [3]. We have also demonstrated the redundancy of some filters across H&E and protein (immunohistochemistry) stained tissues, supporting our basic idea of capturing dominant features across patches and stainings [3].
As next steps we aim to both test the framework on larger data sets and to perform a thorough computational analysis of the different types of cells and discover the patterns that govern cell heterogeneity.
DOLCE is a powerful visual computing technique that enables quantitative description of heavily heterogeneous data and their further computational analysis. DOLCE is further a computational imaging technique that generates quantifiable insights that can help pathologists in their everyday tasks, but also to address the bigger picture; to increase our understanding of physical phenomena, such as pathogenesis, that govern our lives. DOLCE is a great illustrator of how visual computing became a vital component for enabling cognitive medical assistants.
Link:
http://www.zurich.ibm.com/mcs/systems_biology
http://www.wildlab.ch/
References:
[1] E. Zerhouni, et al.: Big Data Takes on Prostate Cancer, ERCIM News, No. 104, January, 2016.
[2] A. Makhzani, B.J. Frey: “A winner-take-all method for training sparse convolutional autoencoders”, CoRR abs/1409.2752, 2014.
[3] E. Zerhouni, et al.: “Disease Grading of Heterogeneous Tissue UsingConvolutional Autoencoder”, SPIE Digital Pathology 2017 (accepted).
Please contact:
Maria Gabrani
IBM Research Zurich











