by Antonino Fiannaca, Massimo La Rosa, Daniele Peri and Riccardo Rizzo
The enormous array of computational techniques and data available due to today’s use of high-throughput technologies can be quite overwhelming for researchers investigating biological problems. For any problem, there are many possible models and algorithms giving different results. We present a new Intelligent System that supports the selection, configuration and operation of strategies and tools in the bioinformatics domain.
The Institute for High Performance Computing and Networking (ICAR-CNR) and the University of Palermo are developing an intelligent system that supports bioinformatics research. The system guides the researcher in building a data analysis workflow and acts as an interface by rendering transparent details regarding the implementation of the tools proposed or the configuration of on-line services.
The system is thus a crossover between classical decision support systems (DSS) and recent workflow management systems (WFMS). It provides both the tools/services needed to resolve a problem, and also the knowledge necessary to justify the choice of specific strategies. This knowledge comprises expertise on the application domain, which is composed of heuristics and strategies derived from bioinformatics literature and experiments and/or provided by one or more human experts.
Architecture
Our system is structured in a three-layer architecture, as shown in Figure 1.
The Interface layer is responsible for interactions between the system and the user. It accepts user queries and returns first suggestions, intermediate and final results. This layer consists of a Graphical User Interface (GUI) and a wrapper module for communication between the GUI and the rest of the system.
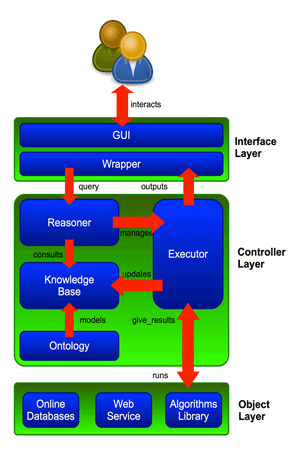
Figure 1: System three-level architecture.
The Controller layer contains the knowledge of the system on the application domain. This knowledge, built on an ontology, contains problem definitions, using pieces of information called facts, and the necessary skills to solve them, using rules in the form IF <precondition is true> THEN <do action>. The controller layer contains the Reasoner component, an inference engine implemented in Java using the Jess engine. The Reasoner makes decisions (inferences) by consulting the knowledge base.
The Executor module is a scheduling agenda whose task is to run the tools and software selected by the Reasoner. The Executor has access to the Object layer (see below), and can update the knowledge base with new facts derived from intermediate results of processing operations.
The Object layer includes information and links to all the instruments the system could use in order to accomplish the user’s request. The components of the Object layer, typically algorithms, web services, online databases and so on, are not part of the system itself: in other words the Object layer offers access to the external tools the system could run.
Decision Making
The decision-making activity of the system is based on an organization of functional modules inside the knowledge base. Each module has its own group of facts and rules, takes care of a specific part of the reasoning process, and is responsible for making decisions about a well defined task. Modules are structured into a hierarchy of levels of reasoning where, at each level, the reasoning is focused on “what to do” and not “how to do”. The highest level is focused on general strategies and directives for solving the user’s request; lower levels in the hierarchy focus on more specialized tasks; the lowest level makes decisions concerning the choice of proper algorithm or service. This hierarchy is set up as a tree, where parent modules manage the activation of children modules when they need a more specific expertise in order to solve the main goal.
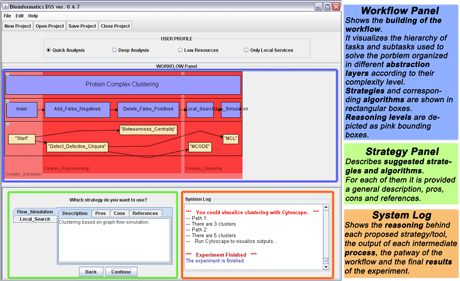
Figure 2: Screenshot of execution of Scenario 1.
Use Cases
We have implemented two simple scenarios to test the system: (1) protein complex extraction from a protein-protein interaction network and (2) reverse engineering gene regularity network.
We collected strategies and heuristics from more than 50 scientific papers and coded them as rules into the knowledge base. These strategies use more than 20 different tools represented in the Object Layer. The system can thus help the researcher to obtain a correct workflow for these scenarios, according to input data and user preferences.
Figure 2 shows a screenshot taken during an execution of the first scenario.
Link:
Jess: http://www.jessrules.com/
Please contact:
Massimo La Rosa
Department of Computer Science, University of Palermo, Italy
Tel: +39 3381918270
E-mail:










