by Magdalena Musielak, Kristian Rother, Tomasz Puton and Janusz M. Bujnicki
Biological functions of many ribonucleic acid (RNA) molecules depend on their three-dimensional (3D) structure, which in turn is encoded in the RNA sequence. We have developed ModeRNA, a program that constructs 3D models of RNAs based on experimentally determined “template” structures of other, related RNAs. This approach is less time and cost intensive than experimental methods.
RNAs are linear polymers comprising tens to thousands of nucleotide residues that function as building blocks. There are four basic building blocks: adenosine, guanosine, cytidine and uridine, but they are often enzymatically modified in the cell to form more than one hundred derivatives with different chemical structures. RNA molecules and their complexes are main players in the production of proteins and other processes in cells. One prominent example is transfer RNA (tRNA), a molecule with a complex 3D structure, used to decipher the genetic code and translate the genetic information in messenger RNA (mRNA) into protein sequences. Determining 3D structures by experimental methods is time-consuming and expensive, compared to methods used for obtaining linear RNA sequences. Thus far only 150 tRNA structures have been solved experimentally, as opposed to more than 300,000 known nucleotide sequences.
Because all tRNAs are evolutionarily related, their structures are similar to one other. We can use experimental 3D data of one tRNA as a template to create a model of another related tRNA with a known, different sequence. In addition to the template structure, information about correspondencies of nucleotide residues between the target sequence and the template sequence (sequence alignment) is required. The alignment is interpreted as a set of instructions regarding which nucleotide residues of the template are to be replaced by which residues of the target. One can think of this concept as creating different pictures from a set of jigsaw puzzle pieces by editing an existing puzzle – for instance replacing a zebra by a lion or adding a mountain in the background in a savannah landscape.
This technique, called homology modelling or comparative modelling, has been implemented in the ModeRNA program. ModeRNA builds RNA structures starting with the easiest part: nucleotides identical between the target and the template are placed in exactly the same position. Then, single nucleotide substitutions are introduced, for example an adenine can be exchanged for a guanine. Finally, parts of the RNA structure that are not present in the template are modelled. For that purpose, ModeRNA uses a library of over 100,000 structural fragments, from which the program chooses the one that geometrically fits best into the insertion site in the model. Because even a well-suited fragment may cause small distortions of bond lengths and angles, ModeRNA can optimize atom coordinates to achieve a stereochemically reasonable conformation. Referring to the puzzle analogy, this would be inserting a set of coherent pieces and applying a rasp to make their edges fit to the remainder of the puzzle.
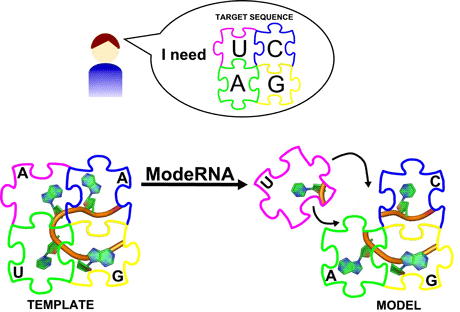
Figure 1: ModeRNA builds a 3D model of an RNA molecule based on a template structure and an alignment of two sequences.
To manipulate 3D coordinates of atoms, ModeRNA employs the Kabsch algorithm for superposition, the Full Cyclic Coordinate Descent algorithm for closing gaps, and the NeRF algorithm to construct atom coordinates. To identify nucleotides, a subgraph matching procedure has been implemented. The program also contains a multitude of functions, eg, to analyse the geometry of existing structures, to find interatomic clashes, or simply remove unwanted nucleotide residues. These functions are available via a scripting interface. ModeRNA has been written in the Python language, using the BioPython library for basic tasks like parsing structural data from the PDB format. ModeRNA is available under the GPL Open Source license. The program is being used by several lab members and collaborators, who provide constant feedback and suggestions for improvement.
To test ModeRNA, we constructed a series of 9801 tRNA models for 100 structures determined by X-ray crystallography. We calculated the root mean square deviation (RMSD) of the atomic coordinates of modelled versus experimental structures. The results showed that the RMSD between the models and the original structures correlates well with the RMSD among experimentally solved structures. The best models reached an RMSD up to 1 Å, with a majority around 4-5 Å. Obviously, the quality of the model depends on how similar the template is to the target, which highlights the importance of the choice of the right template. However, it must be remembered that the RNA structure can change depending on the functional state. For example the anticodon loop and acceptor stem regions of tRNA can adopt different conformations depending on whether the molecule is bound to a protein, or to the ribosome, or if it is free from interactions. We can model these subtle differences by choosing a template structure that is in the desired state.
As the template has such an important influence on model quality, a striking question is: “Do we really need a template, or could we just connect nucleotides from scratch?” In fact, methods like the MC-Fold/MC-Sym pipeline have been used successfully to model small RNA structures (12-50 nucleotides) from nothing more than a nucleotide sequence. However, when the sequence is longer, building a structure without further knowledge becomes computationally unfeasible. For many larger RNA families known 3D structures are available, among them tRNA having around 75 nucleotides, and ribosomes with more than 1000 nucleotides. Thus comparative modeling is a method of choice for 3D structure prediction of large structured RNAs.
In summary, ModeRNA is a tool for construction of 3D models for RNA sequences using structures of another, related RNA as building blocks (templates). ModeRNA also facilitates RNA 3D structure analysis
Link: http://iimcb.genesilico.pl/moderna/
Please contact:
Janusz M. Bujnicki
International Institute of Molecular and Cell Biology, Poland
Tel: +48 22 597 07 50
E-mail:










