by Alexandre Wagemakers and Miguel A. F. Sanjuán
Cells can be considered to be dynamical systems that possess a high level of complexity due to the quantity and range of interactions that occur between their components, particularly between proteins and genes. It is important to acquire an understanding of these interactions, since they are responsible for regulating fundamental cellular processes.
The climax of the genome project, the most notable success of which, has been the complete sequencing of the human genome, was the identification of all the genes that comprise the genetic material of an organism. This achievement led to a new phase of the project: the postgenomic era. Research is now focusing on understanding the organization of, and interactions between proteins, the product of gene expression. Each protein is in charge of a function which can induce changes in other molecules in the cell, such as enzymes or even hormones. These molecules can be viewed as the nodes of a network where the interactions are the links. Thus we can view the system as a complex network of regulation interaction which is responsible for the functioning of the cell.
Recently, the design and the construction of artificial networks has been proposed as a means of studying biological processes, such as oscillations of the metabolism. These networks, simpler than the natural ones, can contribute to the understanding of the molecular basis of a specific function. Simple mathematical models can be constructed in order to perform qualitative and numerical analyses, and synthetic genetic networks can even be synthesized in a laboratory. These works, among others, gave birth to the so-called synthetic biology which integrates several scientific fields such as nonlinear dynamics, physics of complex systems and molecular bioengineering. This is a newly emerging field with a strong interdisciplinary component in which the future advances seem very promising.
The paradigmatic examples of synthetic genetic networks are the genetic toggle switch and the represilator. The genetic toggle switch is the combination of two mutually repressing genes forming a bistable system whose state can be changed with an external signal. One can say that this genetic switch has memory, since it remains in its current state until an external inducer acts again. The second paradigmatic system is the repressilator, which is in fact, a genetic oscillator. In this system three repressor genes are placed in a ring, with each repressor inhibiting the production of the following protein with a given delay. This configuration leads to oscillations in the expression of the three proteins.
Our work in this field, in the Nonlinear Dynamics, Chaos and Complex Systems Group at the Universidad Rey Juan Carlos (URJC), consists mainly of the application of nonlinear dynamics techniques to the modelling and simulation of genetic networks. The evolution of the protein concentration of a particular gene can be represented mathematically with a set of ordinary differential equations. Once these equations are defined, we can apply methods from nonlinear dynamics such as phase space analysis, bifurcation diagrams and stability analysis to understand and predict the behavior of any synthetic genetic network. Furthermore these tools are useful for the design and study of laboratory experiments.
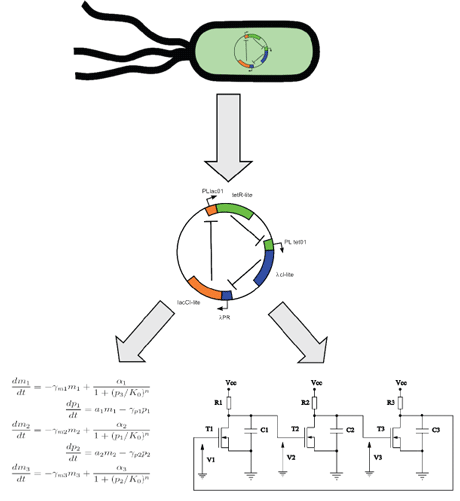
Figure 1: Genetic networks in living cells can firstbe identified with molecular genetics techniques. Once the network is identified a mathematical model is developed and analysed using nonlinear dynamics methods and electronic modelling.
We also have proposed an alternative way to design and analyse the genetic networks with analog electronic circuits. The dynamics of a regulatory genetic network can be simulated with very simple nonlinear circuits based on MOSFET transistors. Our circuits allow a one-to-one correspondence between the structure of the genetic and electronic networks, and their analog character extends this correspondence to the full dynamical behavior. An obvious benefit of this approach is that the electronic circuits are easier to implement experimentally than genetic circuits. We have applied this technique successfully in studies of the dynamics and synchronisation of populations of genetic networks, such as the repressilator and the toggle-switch. Synchronisation of a population of such units has been thoroughly studied, with the aim of comparing the role of global coupling with that of global forcing on the population. We have also analysed a method for the prediction of the synchronisation of a network of electronic repressilators, based on the Kuramoto model. Our research indicates that nonlinear circuits of this type can be helpful in the design and understanding of synthetic genetic networks.
Link:
http://www.fisica.escet.urjc.es/
Please contact:
Alexandre Wagemakers
Universidad Rey Juan Carlos, Madrid, Spain
Tel: +34 91 4888242
E-mail:










