by Christoph M. Friedrich, Christian Ebeling and David Manset
From intracranial aneurysms to paediatric diseases - A biomedical text mining service developed in the European IP-project @neurIST has been integrated into a 3D Knowledge Browser developed in the European IP-project Health-e-Child and can be used for candidate gene searches in different diseases.
Most biomedical knowledge can be found in unstructured form in publications. Every day approximately 2000 new citations are added to the PubMed database, a repository of more than 20 million biomedical citations. Even within specific disease areas it is impossible for scientists to stay up to date. Text mining is seen as a solution to this problem. In the European Integrated-project @neurIST (FP6, 1/2006-4/2010), which was concerned with integrated biomedical informatics for the management of intracranial aneurysms, among other data mining solutions, a biomedical text mining system called SCAIView has been developed. This Knowledge Discovery System, depicted in Figure 1, can answer questions like: “Which genes or gene variations, are concerned with intracranial aneurysms?” Or “Which co-morbidities are mentioned together with Alzheimers disease?” or “Which pathways are involved in diabetes?”.
The key technologies that enable SCAIView are called semantic search and ontology search. The core of the service is a precompiled index that holds a copy of the PubMed database for fulltext searches and additional information on named entities, which occur within the text. A named entity is a semantic entity such as a drug name or gene name which is found by Named Entity Recognition (NER). In the NER process all synonyms of a semantic entity are used for an approximative search and, where possible, the found entity is mapped to a unique database identifier. This process is necessary as some genes or diseases have up to 250 different name variants that occur in text. The information is enriched with data from biomedical databases and ontologies (see Figure 2), so that finally a query for all genes which are on a certain pathway and are co-mentioned with a disease can be submitted.
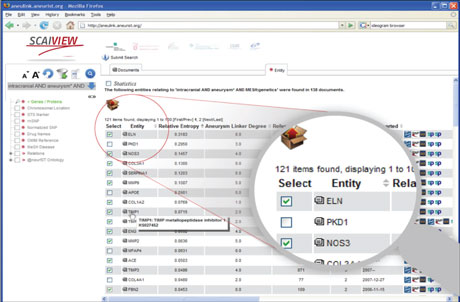
Figure 1: The search interface of SCAIView.
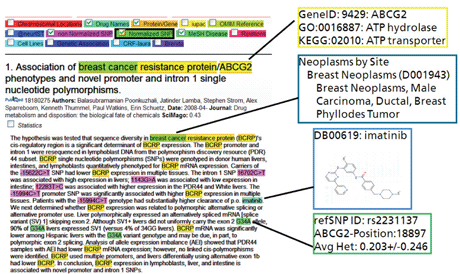 Figure 2: Highlighting and Enrichment through Named Entity Recognition.
Figure 2: Highlighting and Enrichment through Named Entity Recognition.
Most of the named entities in SCAIView are searched with dictionaries but some entities cannot be enumerated beforehand. For these cases rule- and machine-learning based entity recognisers have been implemented. The recognition process involves the machine learning from examples or rules provided by humans. One example for this might be: gene variations like: “... polymorphisms in TIMP-3 (249T>C, 261C>T)”. In this example the result of the search is an identifier to a polymorphism in a database or the identifier of a probe on a Microarray for Genome wide association studies.
Evaluation of Retrieval Performance
Every Search engine is only as good as the results are valid, novel, relevant and useful for the user. The evaluation of SCAIView was based on the question for candidate genes of a disease. The gold standard for “Intracranial Aneurysms” was an expert review on the topic and a Cochrane Report that has been produced in the course of the @neurIST project. We found all candidate genes with our search and they have been distributed among the top ranking hits. Additionally we found other novel candidate genes, which have not been mentioned in the reviews. Other successful evaluations have been conducted for Alzheimer’s disease, schizophrenia and Parkinson’s disease.
Integration into the 3D Knowledge Browser
The integrated European project Health-e-Child (HeC) (FP6, 2/2006-4/2010) has developed a 3D Knowledge Browser, which is specially suited for multi-scale and multi-level searches in biomedical Knowledge Sources. It lacked a search engine for discovering links between HeC data and gene-based information published in external sources. Therefore we conducted a joint research project and integrated the SCAIView results via Webservices into the 3D Knowledge Browser. Now entity searches, for instance for candidate genes, can be displayed next to database searches like a retrieval from SwissProt. In Figure 3 a screenshot of the resulting interface is given. In the Health-e-Child project, this has been used to search for candidate genes in paediatric heart diseases, inflammatory diseases, and brain tumours.
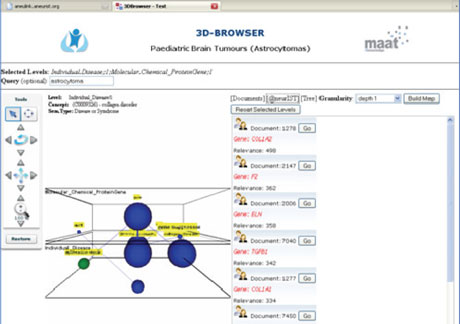
Figure 3: SCAIView searches integrated into the 3D Knowledge Browser.
Links:
http://www.aneurist.org
http://www.health-e-child.org
Please contact:
Christoph M. Friedrich
Fraunhofer-Institute for Algorithms and Scientific Computing (SCAI), Germany
Tel: +49 2241 142502
E-mail:










