by François Fages, Grégory Batt, Elisabetta De Maria, Dragana Jovanovska, Aurélien Rizk
and Sylvain Soliman
The application of programming concepts and tools to the analysis of living processes at the cellular level is a grand challenge, but one that is worth pursuing, as it offers enormous potential to help us understand the complexity of biological systems. To this end, the Biochemical Abstract Machine (Biocham) combines biological principles with formal methods inspired by programming, to act as a modelling platform for Systems Biology. It is being developed by the Contraintes research team at INRIA.
Biologists use diagrams to represent complex systems of interaction between molecular species. These graphical notations encompass two types of information: biochemical reactions (eg, protein complexation, modification, binding to a gene, etc.) and regulations (of a reaction or transcription). Based on these structures, mathematical models can be developed by equipping such molecular interaction networks with kinetic expressions leading to quantitative models. There exist two general categories of quantitative model: ordinary differential equations (ODEs), which offer a continuous interpretation of the kinetics; and continuous-time Markov chains (CTMCs) which provide a stochastic interpretation of the kinetics.
The Systems Biology Markup Language (SBML) uses a syntax of reaction rules with kinetic expressions to define such reaction models in a precise way. Nowadays, an increasing collection of models of various biological processes is available in this format in model repositories, such as www.biomodels.net, and an increasing collection of ODE simulation or analysis software platforms are now compatible with SBML.
Since 2002, we have been investigating the transposition of programming concepts and tools to the analysis of living processes at the cellular level. Our approach relies on a logical paradigm for systems biology which is based on the following definitions:
- biological model = (quantitative) state transition system
- biological properties = temporal logic formulae
- biological validation = model-checking
- model inference = constraint solving.
Our modelling software platform Biocham (http://contraintes.inria.fr/ biocham) is based on this paradigm. An SBML model can be interpreted in Biocham at three abstraction levels:
- the continuous semantics (ODE on molecular concentrations)
- the stochastic semantics (CTMC on numbers of molecules)
- the Boolean semantics (asynchronous Boolean state transitions on the presence/absence of molecules).
Of the three levels, the Boolean semantics is the most abstract. It can be used to analyse large interaction networks without known kinetics. These formal semantics have been related within the framework of abstract interpretation, showing for instance, that the Boolean semantics is an abstraction of the stochastic semantics, ie that all possible stochastic behaviours can be checked in the Boolean semantics, and that if a Boolean behaviour is not possible, it cannot be achieved in the quantitative semantics for any kinetics.
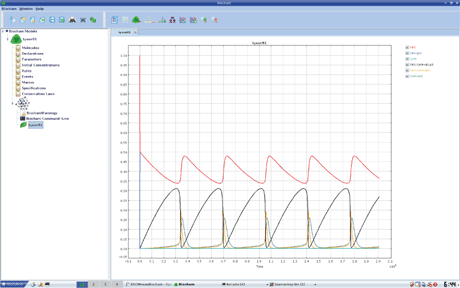
Figure 1: Screenshot of BIOCHAM.
The use of model-checking techniques, developed over the last three decades for the analysis of circuits and programs, is the most original feature of Biocham. The temporal logics used to formalize the properties of the behaviour of the system are respectively the Computation Tree Logic, CTL, for the Boolean semantics, and a quantifier-free Linear Time Logic with linear constraints over the reals, LTL(R), for the quantitative semantics.
Biocham has been used to query large Boolean models of the cell cycle (eg Kohn's map of 500 species and 800 reaction rules) by symbolic model-checking, formalize phenotypes in temporal logic in a much more flexible way than by curve fitting, search parameter values from temporal specifications, measure the robustness of a system with respect to temporal properties, and develop in this way, quantitative models of cell signalling and cell cycle for cancer therapies.
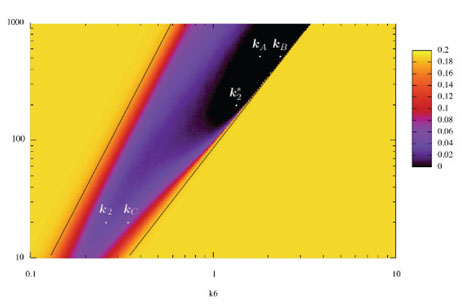
Figure 2: Continuous satisfaction degree of a temporal logic formula for oscillations with a constraint of amplitude in a landscape obtained by varying two parameters.
For some time, an important limitation of this approach was due to the logical nature of temporal logic specifications and their Boolean interpretation by true or false. By generalizing model-checking techniques to temporal logic constraint solving, a continuous degree of satisfaction has been defined for temporal logic formulae, opening up the field of model-checking to optimization in high dimension.
Biocham is freely distributed under the GPL license. In our group, it is currently used in collaboration with biologists at INRA for developing models of mammalian cell signalling, at INSERM and in the European consortium EraSysBio C5Sys for developing models of cell cycle for optimizing cancer chronotherapies, and since 2007 has been used in MIT's iGem competition, for designing synthetic biology circuits.
Link: http://contraintes.inria.fr/Biocham
Please contact:
François Fages
INRIA Paris-Rocquencourt, France
E-mail:










