by Jan Kalina
Gene expression data are typically analysed by standard automated procedures that tend to be vulnerable to outlying values. In a project carried out by the Centre of Biomedical Informatics of the Ministry of Education, Youth and Sports of the Czech Republic, we use alternative approaches, based on robust statistical methods, to measure differential gene expression in cardiovascular patients. Our results are applicable to personalized medicine.
Recent research in the area of molecular bioinformatics and genetics, conducted by the Centre of Biomedical Informatics aims to find the optimal set of genes for diagnostics and prognosis of cardiovascular diseases. Since 2006 we have been using whole-genome beadchip microarray technology to measure gene expression. The sample of peripheral blood is taken from each patient, the ribonucleic acid (RNA) is isolated and applied on the microarray. This technology allows to measure the gene expression as the gene activity leading to synthesis of proteins and consequent biological processes. The Municipal Hospital in Čáslav takes blood samples from two groups of patients: one group with acute myocardial infarct (AMI) or cerebrovascular accident (CVA) (as examples of ischemic diseases); and a control group (patients hospitalized with a different cause without a manifested ischemic disease). This whole-genome analysis examines the entire set of human genes with different microbeads corresponding to different genes randomly distributed on the surface of the microarray, which contains 12 separate physical strips (Figure 1) for samples from different patients.
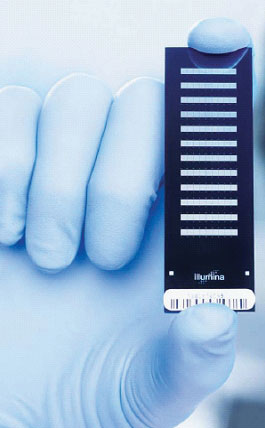
Figure 1: Beadchip for genome-wide expression analysis containing twelve strips.
The standard approach in the preprocessing of the data tends to be vulnerable to outlying observations. The raw data are scanned images with a high fluorescence intensity corresponding to highly expressed genes. To compute the bead-level data for particular microbeads a cascade of transformations is computed, including the local estimation of background, image sharpening and smoothing by averaging, estimating foreground, background correction, data normalization and outlier deletion. The initial steps are strongly influenced by local noise in the neighbourhood of particular microbeads and the resulting biased values are passed on to the next steps of the analysis.The outlier deletion is computed only at the end of the procedure. Therefore the differential expression analysis is sensitive to random or systematic errors in the original data.
As an alternative to the processing of the scanned images with gene expression measurements we propose a more robust approach which involves searching for systematic artifacts in the data. The methods are based on robust statistics applied to image analysis which enables outliers to be deleted at each step of the procedure. The method is also robust to specific properties of the neighbourhood of particular pixels. A careful normalization of bead-level data is computed only after deleting the outlying values. At the same time these methods allow fast computing and are computationally feasible. We propose that standard software for analysing gene expression data be modified to incorporate this new approach.
The outcome of this unique project is the ability to demonstrate which genes are more strongly expressed in patients with acute myocardial infarct or cerebrovascular accident compared to controls. The significance of differential expression of particular genes is acquired by means of statistical hypothesis testing. Clinical and biochemical data recorded for each patient contribute to our understanding of the genetic predisposition to cardiovascular disease. The study of gene expression profiling has allowed the Centre of Biomedical Informatics to patent an oligonucleotide microarray as the main result of the whole study, directly applicable to disease diagnosis, prognosis, prediction and treatment. This technology containing an optimal set of genes provides an invaluable contribution towards the development of a personalized and predictive medical care, in keeping with the new paradigm of data-driven evidence-based medicine.
We believe that the development and use of robust analysis methods is becoming increasingly important in the area of bioinformatics. Next generation (Next-Gen) sequencing, the new low noise approach to genetic analysis, is currently undergoing rapid development, producing huge data sets. Robust statistical methods are therefore becoming ever more crucial for fast and reliable data analysis. It is vital, when designing real-time image analysis systems for Next-Gen technologies, that such methods are adaptive and tailor-made for the particular task, allowing the user to tune parameters. The future of molecular bioinformatics will therefore require more precise and well-considered robust image analysis.
Links:
http://www.euromise.org/cbi/cbi.html
http://www.euromise.org/homepage/department.html
Please contact:
Jan Kalina, Centre of Biomedical Informatics, Institute of Computer Science, Academy of Sciences of the Czech Republic / CRCIM
Tel.: +420 266053099
E-mail:










