Various natural computing paradigms inspired by biological processes, such as artificial neural networks, genetic algorithms, ant colony algorithms, have proved to be very effective and successful. However, we can go one step further: instead of developing computing systems inspired by biological processes, we can now directly use biological substrates and biological processes to encode, store and manipulate information. Early implementations of bio-computing used DNA as a miniaturised "storage medium", but we go beyond this, to harness the "biological nanotechnology" available inside every living cell.
The main objective of the ongoing BACTOCOM project is to build a simple computing device, using bacteria rather than silicon. Microbes may be thought of as biological "micro-machines" that process information about their own state and the world around them. By sensing their environment, certain bacteria are able to move in response to chemical signals, allowing them to seek out food, for example. They can also communicate with other bacteria by leaving chemical trails or by directly exchanging genetic information. We focus on this latter mechanism.
Parts of the internal "program" of a bacterial cell (encoded by its genes and the connections between them) may be "reprogrammed" in order to persuade it to perform human-defined tasks. By introducing artificial "circuits" made up of genetic components, we may add new behaviours or modify existing functionality within the cell. Existing examples of this include a bacterial oscillator, which causes the cells to periodically flash, and cell-based pollution detectors that can spot arsenic in drinking water. The potential for bio-engineering is huge, but the process itself is made difficult by the noisy, "messy" nature of the underlying material. Bacteria are hard to engineer as they rarely conform to the traditional model of a computer or device with well-defined components laid out in a fixed design.
We intend to use the inherent randomness of natural processes to our advantage, by harnessing it as a framework for biological engineering. We start with a large number of simple DNA-based components taken from a well-understood toolbox, which may be pieced together inside the cell to form new genetic programs. A population of bacteria then absorbs these components, which may (or may not) affect the behaviour of the cells. Crucially, the core of our bacterial computer is made up of engineered E. coli. microbes that can detect how well they are performing, according to some external measure, such as how well they can flash in time with light pulses.
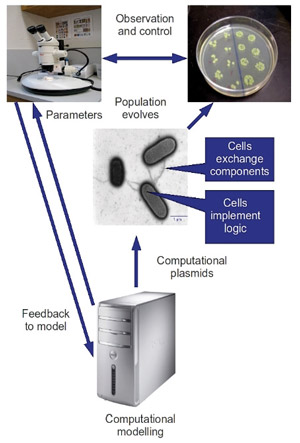
Figure 1: Representation of the structure/function of the BACTOCOM device.
We engineer a set of computational plasmids; circular strands of DNA representing "components", which may be combined together within the cell to form a simple logical circuit. These components may be exchanged between individual bacteria via the process of conjugation; the transfer of genetic material via direct cell-cell contact. By introducing large numbers of these computational plasmids, we initialize the system. Over time, the bacteria integrate the components into their genomes, thus "building" logical circuits. The "output" of these circuits is measurable, and by defining "success" in terms of correlation with a desired signal profile, we allow successful components to flourish via a process of selection. By controlling the desired signal profile, we direct the "evolution" of the population towards novel, robust computational structures.
By performing massively-parallel bacterial random search, we will quickly obtain functional devices without "top down" engineering. There are many potential benefits to this work, from both a biological and computing perspective. By "evolving" new functional structures, we gain insight into biological systems. This, in turn, may suggest new methods for silicon-based computing, in the way that both evolution and the brain have already done. In building these new bio-devices, we offer a new type of programmable, microscopic information processor that will find applications in areas as diverse as environmental sensing and clean-up, medical diagnostics and therapeutics, energy and security.
The BACTOCOM project is a three-year programme, funded by the European Commission FP7 FET Proactive initiative: CHEM-IT (Bio-chemistry-based Information Technology). It began in February 2010, and is coordinated by Martyn Amos of Manchester Metropolitan University, UK. Other partners include, Universite d'Evry Val d'Essonne-Genopole®-CNRS (France), Universidad Politécnica de Madrid and University of Cantabria (Spain), Interuniversitair Micro-Electronika Centrum (IMEC, Belgium) and Charite - Universitätsmedizin Berlin and the Technical University Munich (Germany).
Link:
BACTOCOM project website: http://www.bactocom.eu
Please contact:
Martyn Amos
Novel Computation Group, Manchester Metropolitan University
E-mail:










