by Olivier Colliot (CNRS)
In artificial intelligence for medicine, more interpretable and reliable systems are needed. Here, we report on recent advances toward these aims in the field of brain diseases.
As in different fields of medicine, AI holds great promise to assist clinicians in the management of neurological diseases. However, there is still an important gap to bridge between the design of such systems and their use in clinical routine. Two major components of this gap are interpretability and reliability.
“Interpretability”, the user’s ability to understand the output provided by an AI system, is important for the adoption of AI solutions by clinicians. To make AI systems more interpretable, different lines of work are being pursued. A first avenue is to explain the basis of a prediction based on the input features. While this is relatively straightforward for linear models, it is more difficult for complex non-linear techniques such as deep neural networks, even though advances have recently been made in this area.
Another complementary way to make AI systems more interpretable is to predict not only a clinical outcome (e.g. diseased/healthy, lesional/non-lesional) but also different types of medical data and measurements characterising a patient. For instance, in Alzheimer’s disease, one may try not only to predict the future occurrence of dementia, but also the future value of cognitive scores or future medical images of the patient. Recently, we proposed a system to predict brain images that are representative of different pathological characteristics in multiple sclerosis. In a first work [1], we designed a system that can predict a specific type of magnetic resonance image (MRI), called FLAIR, from other types of MR images. We showed that overall the predicted images did a good job of preserving the characteristics of the original image. We then applied an automatic segmentation algorithm and showed that its results on the predicted and original images are consistent.
We further proposed an approach to predict myelin content from multiple MRI modalities (Figure 1) [2]. Myelin is a substance that wraps axons and increases the speed of transmission of information between neurons. Multiple sclerosis is characterised by the loss of myelin (demyelination) whose quantification is essential for tracking disease progression and assessing the effect of treatments. Myelin can be measured in vivo using positron emission tomography (PET) with specific tracers. However, PET is an expensive imaging modality and is not available in most centres. We showed that it is possible to synthesise PET images from multiple MR images, which are less expensive to acquire. The predicted image allows accurate quantification the amount and location of demyelinated areas. These results will need to be confirmed on larger, multicentric, datasets. By providing both the quantified outcome (demyelination) and the predicted image, our approach has the potential to be more interpretable for the clinician.
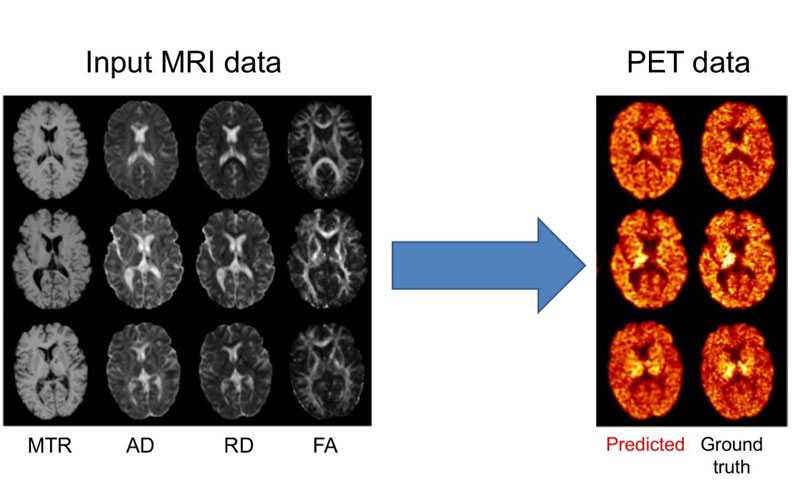
Figure 1: Prediction of myelin content, as defined from PET images, using multiple MRI modalities. On the left, input MRI modalities: magnetisation transfer ratio (MTR) and three measures computed from diffusion MRI, axial diffusivity (AD), radial diffusivity (RD) and fractional anisotropy (FA). On the right: predicted and ground truth PET data. The PET tracer is the Pittsburgh compound B (PiB) is used to measure myelin content in the white matter of the brain.
Naturally, reliability is a mandatory property of medical AI systems. Design of reliable systems involves many steps from evaluation of prototypes to certification of products. At the stage of academic research, an important component is the ability to replicate results of a given study. Replication is indeed a cornerstone of scientific progress in all areas of science. Such a process involves two related but distinct aspects: reproducibility, defined as the ability to reproduce results based on the same data, and replicability, the reproduction using different data.
In a recent work, we studied the problem of reproducibility of deep learning approaches for assisting diagnosis of Alzheimer’s disease from MRI data [3]. First, we reviewed existing studies and unveiled the existence of questionable practices in a substantial number of them. Specifically, among the 32 studies, half of them potentially introduced data leakage (the use of some information from the test set during training). Data leakage was clear in six of them and possibly present in 10 others which had insufficient details about their validation procedure. This is a serious problem, particularly considering that all these studies have undergone peer-review. Given these defects in the validation procedure, it is unlikely that the very high performances reported in these studies would be replicated by others. Moreover, many of these works were not reproducible because the code and data were not made available.
We thus proposed a framework for reproducible experiments on machine learning for computer-aided diagnosis of AD [3]. The framework was composed of standardised data management tools for public data, image preprocessing pipelines, machine learning models and validation procedure. It is open-source and available on github [L1]. We applied the framework to compare the performance of different convolutional neural networks and provide a baseline to which future works can be compared. We hope that this work will be useful to other researchers and pave the way to reproduce research in the field.
Link:
[L1] https://github.com/aramis-lab/AD-ML
References:
[1] W. Wei, E. Poirion, B. Bodini, S. Durrleman, O. Colliot, B. Stankoff, N. Ayache: “Fluid-attenuated inversion recovery MRI synthesis from multisequence MRI using three-dimensional fully convolutional networks for multiple sclerosis”, J Med Imaging 6:27, 2019. DOI: 10.1117/1.JMI.6.1.014005, 2019
[2] W. Wei, E. Poirion, B. Bodini, S. Durrleman, N. Ayache, B. Stankoff, O. Colliot: “Learning Myelin Content in Multiple Sclerosis from Multimodal MRI through Adversarial Training, in: Proc. MICCAI - Medical Image Computing and Computer Assisted Intervention, Springer, 2018.
[3] J. Wen, E. Thibeau-Sutre, J. Samper-Gonzalez, A. Routier, S. Bottani, S. Durrleman, N. Burgos, O. Colliot: “Convolutional Neural Networks for Classification of Alzheimer’s Disease: Overview and Reproducible Evaluation”, 2019. ArXiv190407773 Cs Eess Stat.
Please contact:
Olivier Colliot
CNRS, Inria, Inserm, Sorbonne University, Brain and Spine Institute Paris, France










