by Kostas Stefanidis, Giorgos Flouris, Ioannis Chrysakis and Yannis Roussakis, ICS-FORTH
D2V, a research prototype for analysing the dynamics of Linked Open Data, has been used to study the evolution of biomedical datasets, such as the Experimental Factor Ontology (EFO) and the Gene Ontology (GO).
Datasets are continuously evolving over time as our knowledge increases. Biomedical datasets in particular have undergone rapid changes in recent years, making it difficult for engineers and scientists to follow the evolution and keep up with recent developments.
To address the problem of managing evolving datasets and understanding their evolution, we have developed D2V [L1], a research prototype designed for studying the dynamics of web data and the associated Linked Open Data (LOD). Specifically, D2V is able to detect and analyse changes and the evolution history of LOD datasets, thereby allowing remote users of a dataset to identify changes, even if they have no access to the actual change process. Interestingly, it also empowers users to perform sophisticated analysis on the evolution data, so as to understand how datasets (or parts of them) evolve, and how this evolution is related to the data itself. For instance, one may be interested in specific types of evolution, e.g., classes becoming obsolete, or in the evolution of a specific entity (e.g., a specific disease or genome). Our tool aims to become a critical addition to the arsenal of data analysts and scientists for dynamicity analysis in biomedical or other datasets.
For example, consider Alice, a specialist in the field of genetic diseases, who is interested in the connections between human genome and diseases, and frequently annotates the Disease Ontology [L2] with her research findings. As ontology terms change, often becoming obsolete, i.e., replaced by other terms, Alice needs an easy way to identify the changes related to the object of her study and understand how these changes affect her research. To help Alice, D2V allows her to manage, detect, and view changes in a variety of ways.
In particular, D2V handles two types of changes, with the aim of making changes intuitive and human-understandable. The first is simple changes, which are fine-grained changes defined at design time and provide formal guarantees on the soundness and completeness of the detection process. The second type is complex changes, which are custom-built and defined at run-time by the user to satisfy application-specific needs; for example, complex changes may be used to report coarse-grained changes, changes that are important for the specific application or user, changes with special semantics, or changes that should not happen at all (their detection being like an alert for an abnormal situation).
To detect changes, we rely on the execution of appropriately defined SPARQL queries, a W3C standard for querying LOD. The answers to these queries determine the detected changes, which are represented as instances of an ontology of changes (Figure 1). This allows the connection of the detected changes with the actual data using standard LOD principles, blending the data with the evolution history and supporting navigation among versions, cross-snapshot and historic queries. The analysis of the evolution history is based on SPARQL.
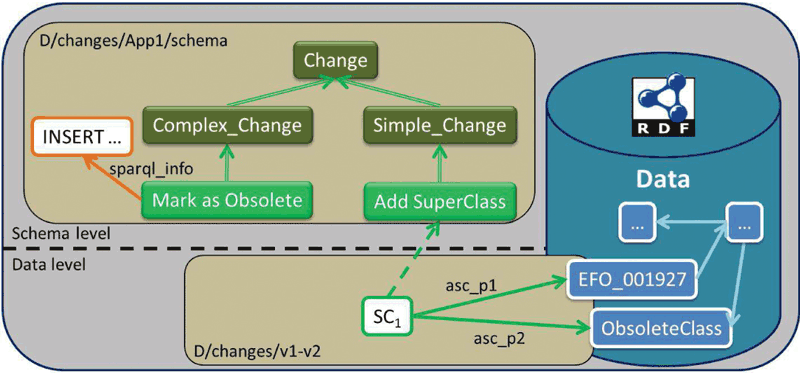
Figure 1: The ontology of changes.
The retrieval of the detected changes from the ontology allows them to be presented to the user, through interactive interfaces and visualization paradigms (see Figure 2 for a visualization of EFO [L3] evolution). We provide different views of the evolution history for different types of analyses: the user can see the evolution history of a given URI (term-centric view – e.g., return all changes associated with “adrenal gland disease”), of the dataset as a whole (dataset-centric view – e.g., view all changes in the Disease Ontology), or of specific versions (version-centric view – e.g., view all changes between a given pair of versions); or the user may be interested in a change-centric view, where the instantiations of a given change are reported (e.g., return all classes that were made obsolete); the user can filter the different results to a fixed set of changes, change types, or versions; or visualize evolution along a series of consecutive versions, or for an arbitrary pair only.
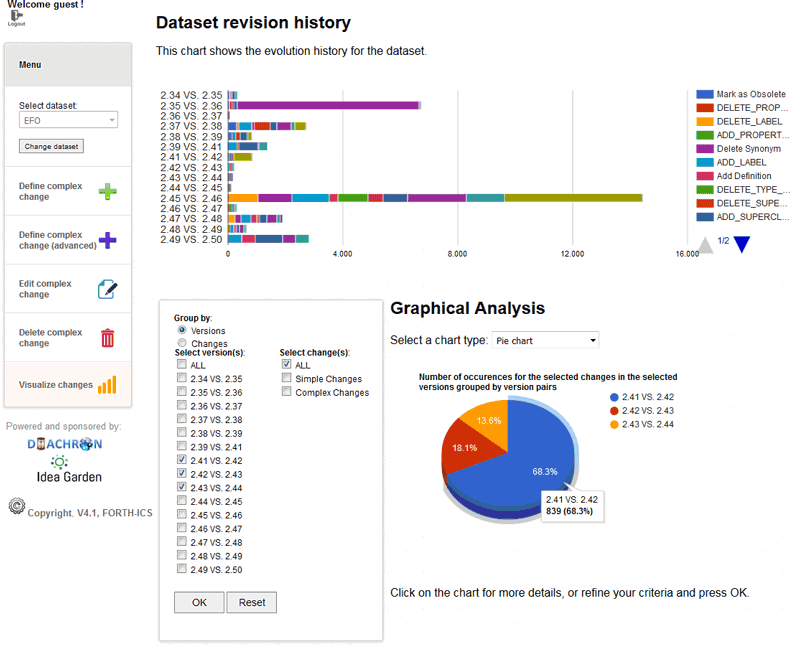
Figure 2: Dataset-centric and version-centric view.
We designed and developed D2V at FORTH-ICS, an ERCIM institute, in collaboration with researchers from ATHENA and EMBL-EBI, in 2014-2015. We plan to enhance our software with additional features, and incorporate additional biomedical datasets.
The development of D2V was funded by the EU FP7 projects DIACHRON and IdeaGarden. Further details can be found at [1], [2].
Links:
[L1] http://www.ics.forth.gr/isl/D2VSystem/
[L2] http://disease-ontology.org/
[L3] http://www.ebi.ac.uk/efo/
References:
[1] Y. Roussakis, et al.: “A Flexible Framework for Understanding the Dynamics of Evolving RDF Datasets”, Best Student Paper Award, ISWC-15, Research Track.
[2] Y. Roussakis, et al.: “D2V: A Tool for Defining, Detecting and Visualizing Changes on the Data Web”, ISWC-15, Demonstrations Track.
Please contact:
Kostas Stefanidis, ICS-FORTH, Greece
Tel: +30 2810 391635











