Digital Biological Cell
by Tomá Bílý and Michal Karásek
Can mathematicians describe digital cells in the same way they can mimic the behaviour of biological systems? It appears that digital cells can mimic some behavioural properties, while others are being intensively studied. From some level of approximation, we can use this knowledge to connect the predictive power of these digital cell models to the methods commonly used in clinical general practice.
When we refer to a digital biological object, we usually mean a mathematical or computer model of a real object. We shall use the adjective "digital" in this sense. The digitization of the biological cells of digital patients and organisms is the main paradigm of theoretical biology and medicine. It is a crucial instrument for predicting the behaviour of biological systems over scales ranging from nanometres to the whole organism. Researchers at the "Seminar on Mathematical and Computer Predictions of Cell Behaviour", which has been held for over a decade at the Department of Applied Mathematics (KAM) at Charles University, Prague, Czech Republic, are working on a digital cell project. There are two main streams of research. In the first, we invent mathematical models of a universal digital cell that can mimic certain important aspects of eukaryotic biological cells. In the second, we study these models collaboratively using interactive well-behaved simulation programs called virtual laboratories.
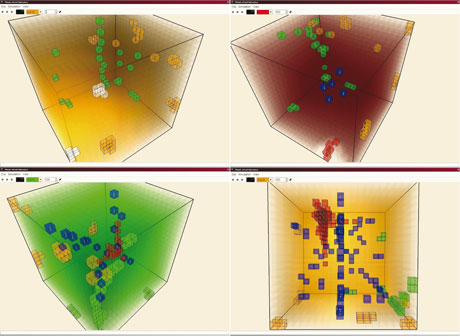
From a mathematical point of view, we are using multiscale hybrid continuous-discrete models; these are based on reaction-diffusion models ruled by partial differential equations on the finest hierarchical level (small spatial scales and short time intervals), and discrete agent-based models with universal rules (some based on metabolic control ruling) on coarser levels. Our actual research target is to produce qualitative models and understand their combinatorial properties.
A virtual laboratory is a special simulation program with the following characteristics. It is designed as an implementation of a mathematical model and its behaviour is restricted by the validity of the model rules. The actions of model entities can be modified by changing the parameters of its environment and/or the entities interaction rules. Although realistic behaviour is not fundamental, parameterization helps in looking for biological plausibility. In addition, incremental parametrical changes allow researchers to identify important stable and dividing situations. For consistency in simulation it is determined that virtual experiments are always (ceteris paribus) repeatable with the same results. Visualization of the virtual laboratory output is absolutely necessary.
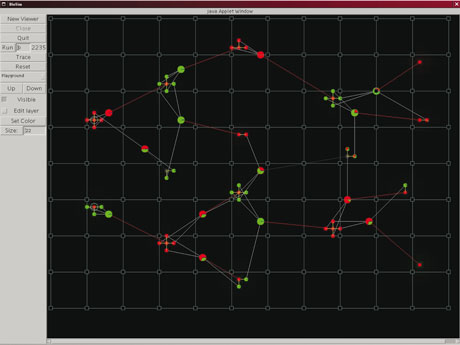
Moreover, the concept of the virtual laboratory is a natural way to explain an abstract experiment in an interpersonal communication. Using a number of prepared experiments allows a better understanding of our digital cell models, and facilitates both general identification with problems and model behaviour orientation. According to our experience, using a visual form of information is crucial for interdisciplinary communication.
Our virtual laboratory design makes it possible for an experimenter to change model parameters easily, which with adequate visualization also enables the use of the laboratory as an educational tool. Either alone or under supervision, users can modify the parameters of an experiment and follow the corresponding changes in simulation behaviour. Collaboration in teams is supported and recommended.
We have implemented ligand-based models of neural network growth dynamics, immune cell dynamics (eg allergic reactions or different blood-type reactions) and the morphogenetic formation of shapes. We have compared the results of simulations with biological experiments and it seems that certain qualitative properties of the biological experiments can be successfully simulated.
Several objectives will be addressed in our future work. First, the modularization of our virtual laboratory system will be improved. Second, we will run larger virtual experiments in order to simulate some simple digital organisms and thus explore more combinatorial properties. Finally, we will perform more quantitative virtual experiments and then get it closer to clinical general practice.
Please contact:
Tomá Bílý
Charles University, KAM MFF - CRCIM
Tel: +420 221914261
E-mail: tomby![]() kam.mff.cuni.cz
kam.mff.cuni.cz










