by Mike Holcombe
Systems biology, an integrated research field involving experimental biology and computational modelling, takes a systems-level view of biological phenomena without losing the detail and complexity that is inherent in all biological systems. I call this the 'in virtuo' approach (in preference to 'in silico' which seems to imply a specific computational technology). Two investigations are discussed: one looking at how part of the innate immune system works, and the other at how skin seems to heal wounds.
Our key approach to modelling in this area is to start at the bottom and work up. This is in contrast to more traditional approaches that are based on the use of differential equations of various types and which take a top-down approach. Biological systems are composed of many different types of coherent components that communicate with each other in a variety of ways, and develop structures and functions as emergent phenomena. The interesting thing about these systems is that there is no overall control centre; that is, the systems organize themselves according to their hard-wired instructions and the laws of physics and geometry.
One factor with this type of modelling, known as agent-based or individual-based modelling, is that a massive number of components must be modelled "often into the millions" and this requires significant computational resources. Each component is designed as a software "agent" with its own life cycle, functional behaviour and structural form. These agents communicate with one another through many different communication channels; these may be chemical signals, physical contact and so on. This approach views each agent as a communicating "X-machine" a fully general model with many attractive and convenient properties. We try to model the agents as accurately as possible using all the experimental techniques of molecular biology, and to then put them all together in simulations. We continuously track the position of every agent, since where it is and what it is near has a fundamental effect on what it does. For example, if we are modelling at the cellular level and a particular cell is ready to divide, it cannot do so if it is surrounded by other cells and there is no space into which it can "expand". Validation must be done experimentally in a continuous manner.
We have developed a framework FLAME (Flexible Agent-based Modelling Environment) that allows for the simple definition of the agents and their environments, and compiles straight to highly optimized code for running on parallel computers.
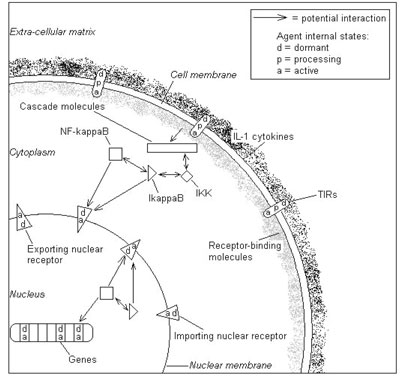
The first example is the NF-kB system a vital part of the innate immune system. In this system the NF-kB molecules are bound to other molecules such as IkBa and the onset of infection causes them to separate and the NF-kB molecules then move into the nucleus and switch on a number of key genes. The agents in this model are varied and in some cases short-lived. For a biochemical pathway, this means that anything from a molecule to a signalling receptor to an entire chain of interactions can be modelled as an agent, thus providing a modular and extensible modelling framework that allows abstraction of detail as necessary.
In this model, molecular agents diffuse through the cell, binding and dissociating from other molecules, receptors and cell structures in accord with signals they send and receive from surrounding agents. Every agent is represented by a complete computational model, the communicating stream X-machine. This provides an intuitive and rigorous basis on which to model the functional behaviour of systems in a flexible and extensible manner. An important feature is the memory of each agents X-machine, which contains its physical location, meaning that the number of states required to model the system is manageably small. It is essential that the agents are both biologically plausible as entities and that their behaviour is based on experimental measurements. In the model, as in reality, molecular interactions are local events that depend only on the position and current state of the molecules involved, where the state of a molecule is whether or not it is already bound. The physics of a molecule is modelled according to specific agent-based characteristics, including which types of interaction are possible. If two molecules may interact according to the rules, they must satisfy criteria on their state and proximity, derived from standard rate constants. If interaction occurs, the state of each agent changes to a "bound" state, which can be reversed through random thermal separation.
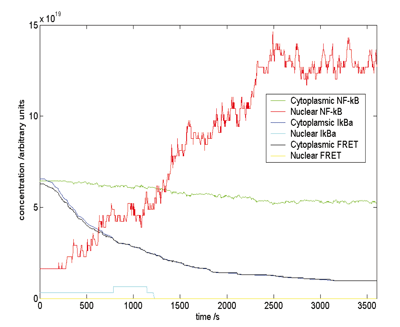
Not only is this the most detailed and accurate model of this pathway so far, but it has led to a new biological discovery. There had been some evidence that the ratio of one of the key molecules IkBa to NF-kB was three times what was "needed". Where was all this excess IkBa? The model predicted that it could be sequestered with actin filaments, and recent experiments have produced significant data that confirms this.
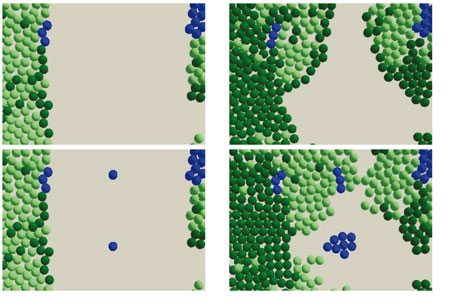
The second case study involves the process of skin healing. The Epitheliome project is part of the Human Physiome Project, and aims to integrate computational and biological models of the social behaviour of cells within epithelial tissue. We aim to develop a computational model of cell behaviour within the context of tissue architecture, differentiation, wound repair and malignancy.
Factors in the model include cell division (agent proliferation) and cell death; physical space (the cells cannot divide if there is no room); intracell communication; cell role changes according to age and circumstance; and other variables like nutrient uptake, immune response, calcium and physical force.
We have investigated the simulation and the tissue culture of stem cells and how they develop into tissue. The simulations have demonstrated that the distribution of the Keratinocyte stem cells determines to a large extent how successful the repair will be. For instance, new cells will fail to spread if there are large gaps between them, because the growth factor signal from the stem cells is too dilute.
Link:
FLAME (Flexible Agent-based Modelling Environment): http://www.flame.ac.uk
Please contact:
Mike Holcombe
University of Sheffield, UK
E-mail: m.holcombe![]() dcs.shef.ac.uk
dcs.shef.ac.uk










