by Hervé Delingette, Maxime Sermesant, Nicholas Ayache, Dominique Chapelle, Miguel Fernandez, Jean-Fréderic Gerbeau and Michel Sorine
The CardioSense3D action is an INRIA initiative that aims to develop a patient-specific simulation of cardiac activity that is suitable for clinical applications. This simulation includes a coupled model of the electrophysiological and mechanical behaviour of the heart, whose parameters are estimated based on the medical images and signals acquired on a given patient.
There is an irreversible evolution of medical practice toward more quantitative and personalized decision procedures for prevention, diagnosis and therapy, based on ever larger and more complex sets of measurements. This deep trend has led to a crucial need to produce a new type of so-called computational model of the anatomy and the physiology of the human body, which is able to explain the observations, detect abnormalities, predict evolutions, and to simulate and evaluate therapies.
The simulation of the heart is the subject of growing attention due to the impact of cardiovascular diseases in industrialized nations and to the high complexity of the cardiac function. Indeed, formulating a computational model of the cardiac function of a specific patient represents a great challenge due to the intrinsic physiological complexity of the underlying phenomena, which combine tissue mechanics, fluid dynamics, electrophysiology, energetic metabolism and cardiovascular regulation. Another source of difficulties lies in the partial information on cardiac function available for a specific patient through acquired signals and images.

CardioSense3D
To tackle these challenges, a four-year Large Initiative Action was launched in 2005 and funded by the French national research centre INRIA, focusing on the electromechanical modelling of the heart. This action relies on the expertise of four INRIA research teams (Asclepios, Reo, Macs and Sisyphe, previously known as SOSSO2) covering the fields of medical image analysis, computational structural and fluid dynamics, numerical analysis and control. It is also a collaborative framework that involves clinical centres such as the Guy's Hospital London, the Laboratory of Cardio-Energetics at the National Institutes of Health and the Hospital Henri Mondor. These and other partners are listed on the Web site of the project.
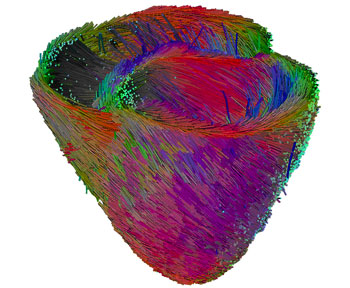
A prime objective of the CardioSense3D project is to build a personalized cardiac simulator with identifiable parameters, which links four different physiological phenomena: electrophysiology, mechanical contraction and relaxation, myocardium perfusion and cardiac metabolism. In building patient-specific cardiac models, the complexity of these models is constrained to match that of the available observations. Furthermore, an important research activity in the CardioSense3D action lies in the conception of data assimilation software that can estimate patient-specific parameters and state variables from given signals or images of the cardiac activity.
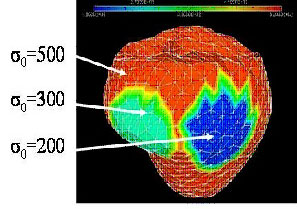
The current CardioSense3D model includes a simulation of the electrical depolarization and repolarization of the cardiac tissues through a set of macroscopic reaction-diffusion equations. This electrical activity can be synchronized with the actual ECG (electrocardiogram) of the patient, and creates a mechanical contraction and an active relaxation that are modelled by a set of partial differential equations.
We have shown that this model can be interactively adjusted to the actual geometrical, mechanical or electrical properties of a patient's heart through the use of conventional or tagged magnetic resonance images and some in vivo electrophysiological measurements. The average direction of the myocardium fibres is also integrated into this model, because it plays an important role in both the electrical and mechanical modelling. Even more interestingly, the model can then be used to study the effect of locally modifying some electrical or mechanical properties in order to better predict the effect of a therapy or the evolution of a pathology.
We believe that this kind of model of a dynamic organ could be used in the future to better plan or train a number of medical gestures; for instance, in radio-frequency ablation procedures for the positioning of pacemaker stimulation probes.
Link:
http://www.inria.fr/CardioSense3D
Please contact:
Hervé Delingette
INRIA, France
Tel: +33 4 92 38 76 60
E-mail: Herve.Delingette![]() sophia.inria.fr
sophia.inria.fr










