For the clinician, predicting fracture risk for individual patients essentially amounts to the quantitative analysis of bone density. However, several studies have shown that bone strength, an indicator for bone fracture risk, is only moderately predicted by bone density. This is not surprising: bones are not solid structures, but are made up of an outer shell of compact bone enclosing a core of spongy, trabecular bone. This trabecular bone, which is located at the end of long bones, has a porous structure that contributes significantly to the load-bearing capacity of the human skeleton. It is not a random structure; rather its trabeculae are typically aligned with the main loading direction.
The investigation of the mechanical properties of trabecular bone presents a major challenge. This is due to its high porosity and complex architecture, both of which vary substantially between anatomic sites and across individuals. A promising technique that takes bone microarchitecture into account is microstructural finite element (microFE) analysis. A very large number of finite elements is needed to accurately represent a human bone with its intricate microarchitecture; hence, the resulting microFE models possess a very large number of degrees of freedom.
Detailed microFE models are obtained through high-resolution micro-computed tomography (microCT) of trabecular bone specimens. This allows nondestructive imaging of the trabecular microstructure in living patients with resolutions on the order of 80 microns.
The discrete formulation is based on a standard finite element (voxel) discretization for linear elasticity. The degrees of freedom represent the displacements in the three axis directions at the voxel vertices.

A typical problem size is 100 million voxels corresponding to about 300 million degrees of freedom. Higher resolutions or larger bone specimens lead to larger models. We have recently simulated bones with up to 1.6 billion degrees of freedom (see Figure 1). A problem of 1 billion degrees of freedom generates a data volume essentially the system matrix of more than 1.25 TB, which must be stored in main memory.
We solve the resulting symmetric positive definite linear systems by the preconditioned conjugate gradient algorithm. We employ an algebraic multigrid (AMG) preconditioner based on smoothed aggregation. A particular strength of our parallel finite element solver, ParFE, is the ability to construct the preconditioner without actually forming the system matrix. This matrix-free procedure reduces the aforementioned memory requirements by 75%, meaning that only a quarter of the processing elements is needed to solve the problem. Our rule of thumb is half a million degrees of freedom per gigabyte of main memory. We use ParFE on the Cray XT3/XT4 and on the IBM Blue Gene to solve real-world problems, with the typical time to solution being just a few minutes. In contrast to pure bone computations, implants entail variable material parameters. Bone grafts and biomaterials are often used to aid the repair of complicated fractures. The healing process can be analysed using recently developed high-resolution in vivo peripheral quantitative computed tomography (pQCT). While pQCT is able to provide detailed information on bone structure, it cannot do the same for mechanical stability. However, such information can be derived from microFE analyses, and our solver can handle this multi-material situation without problems. Stiffness values as assessed from microFE analyses correlated well with the experimentally measured stiffness.
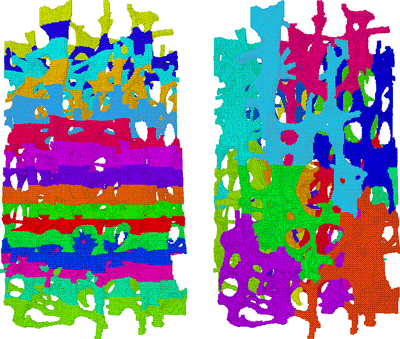
A crucial issue in large-scale computations is load balancing. This holds true in particular for complicated structures such as trabecular bones. The layered data layout that is induced by the CT data is not appropriate for the structural analysis, but must be redistributed in order to reduce interprocessor communication (see Figure 2). This portion of the code, that is the repartitioning, does not yet scale beyond 1000 processors. We need to resolve this issue before we can efficiently tackle problem sizes of several billion degrees of freedom. We are also working on an entirely new approach that exploits the underlying Cartesian grid in order to save more memory space.
A further important issue is the visualization of the results. This work is done interactively and hence takes much more wall clock time than the simulation itself. The large data volume again requires parallel processing.
This project is a collaboration between the Institute of Computational Science and the Institute for Biomechanics (both at ETH Zürich), the Swiss Supercomputing Centre (CSCS) and the IBM Zurich Research Center.
Links:
http://people.inf.ethz.ch/arbenz/projects/bone.html
http://parfe.sourceforge.net
Please contact:
Peter Arbenz
Institute of Computational Science, ETH Zürich
Tel: +41 44 632 74 32
E-mail: arbenz![]() inf.ethz.ch
inf.ethz.ch
Ralph Müller
Institut for Biomechanics, ETH Zürich
Tel: +41 44 632 45 92
E-mail: ram![]() ethz.ch
ethz.ch










