In silico simulation methods - simulations within computer software - have become indispensable tools in the development of expensive new technology, from aircraft manufacture to nuclear power station development. For various reasons however, modelling and simulation techniques have only very slowly been adopted by the biological research community.
Biological systems are typically complex and adaptive. Given the dynamic nature of these phenomena, it is non-trivial to provide a comprehensive description of such systems and, in particular, to define the importance and contribution of low-level unsupervised interactions to the overall evolution process. An agent-based approach is presented here.
The immune response, in particular, consists of relatively simple cell-level interactions, but is capable of highly diverse and individual behaviour, with outcomes that are self-evidently more than the sum of their parts. In what follows, a one-to-one reciprocity between autonomous agents and immune cells is proposed. The emergence of tissue-level and body-level patterns is clearly dependent on the relative sizes of cell populations and the balance these achieve. This is demonstrated, for example, by the course of HIV progression, where the whole immune system collapses once immune cell counts decrease below critical levels.
High temporal granularity is necessary to realistically account for cell mobility and interactions. With current computing resources, this prevents models accounting for every immune cell of every type over the whole body. A compromise between agent diversity and agent population size is therefore required. We have implemented a flexible model, which currently includes viral agents and an agent type for each entity of the cell-mediated response. This guarantees enough diversity to account for principal immune and viral mechanisms, while permitting the simulation of large populations.
In this way, cell mobility can be examined. Agents are explicitly located in structures modelling lymph nodes. Agent movement within these leads to the occurrence of cell-level interactions, and their passage from one node to another. Thus, lymphatic chain structures allow for the spread of viral infection and connected immune response. The model successfully accounts for rapid spreading throughout the initial lymph chain (ie within hours), and slower progression through the whole body (weeks being necessary to obtain infection of all nodes).
Using parallel implementation on a large cluster computer, simulations can model up to a thousand lymph nodes for a total of more than one billion agents. This permits accurate modelling of the immune response to infections, and inclusion of localized effects such as HIV early infection within the gastrointestinal tract, whose importance was recently highlighted. Equivalent large-scale frameworks are also applicable to other complex systems, biological or otherwise. In particular, in silico techniques have enormous potential to contribute to the quality and cost-effectiveness of the drug development process.
Visualization of the results from such simulations is non-trivial. In what follows, we present some initial results in the visualization of immune responses that allows the user to parameterize their in silico experiments in order to study the maximum infection clearance dynamics. The tool also proves to be a useful aid in the classroom for students in their early undergraduate training.
Collaboration between ITT Dublin and Dublin City University has led to the development of novel visualization techniques to study cellular interaction from both the large-scale spatial and temporal perspectives. Visualization of processes offers the researcher the ability to speed up, slow down and hypothesize various parameters. Visualization aids understanding as we rely on visual perception to make crucial decisions. For example, with our initial model, we can visualize the dynamics of an idealized lymphatic compartment, with APC and CD8 cells.
We can parameterize many aspects, such as initial cell levels, rates of change in the life cycle, frequency of infection events and many others. Some viral pathogens are capable of persistent re-infection in that, although population levels of infected antigen presentation cells may decline in response to clearance pressure by a specific CD8 response, over time the number of infected cells rises to chronic and sometimes acute levels. Examples of such viruses are HIV, Human T-lymphotropic virus (HTLV) hepatitis C (HCV), hepatitis B, cytomegalovirus CMV, Epstein-Barr virus (EBV) and rubella.
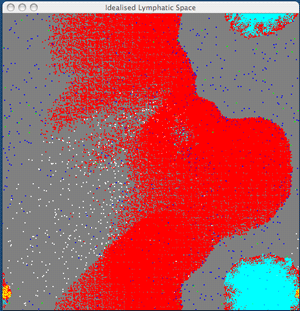
From the figure we see that as the primary response continues, the effector cells now begin their exit from the lymphatic compartment, after which they will be carried through the blood system and will migrate towards the site of the initial infection (if one exists).
We have developed a 'front end' visualization component to allow students and lecturers in the classroom and lab to experiment with a variety of parameters. We are strongly motivated in this research by the findings of a recent EU report indicating that in silico modelling and simulation of biological processes is a key requirement in the development of cost-effective and timely new disease therapies.
Link:
http://sci-sym.computing.dcu.ie
Please contact:
Dimitri Perrin
Dublin City University, Ireland
Tel: +353 1 700 8449
E-mail: dperrin![]() computing.dcu.ie
computing.dcu.ie
John Burns
Institute of Technology Tallaght, Ireland
Tel: +353 1 404 2766
E-mail: john.burns![]() ittdublin.ie
ittdublin.ie









