Recent research shows that the role of the epigenetic events in the development of cancer is more prevalent than previously assumed. An important need has been identified to integrate epigenetic pathways in the management of gastro-intestinal malignancies (A. Rachid, J.P.J. Issa, Gastroenterology, 127, 1578-1588, 2004).
In the context of cancer, recent work dealing with epigenetics in the study of colorectal cancer has shown remarkable improvements in experimental quantitative identification of epigenetic events. For example, [Ogino 2006] have published results which suggest that methylation of the promoter of a gene is not merely a binary value as previously supposed, but that intermediate degrees of methylation can be determined, which influence the extent of the gene expression. Linking results such as this with eg the stage of cancer development, the cell characteristics at this stage and other factors, should provide considerable insight on the dynamics of cancer development. The experimental technique described by S. Ogino, M. Brahmandam et al, Modern Pathology, 19, 1083-1090, 2006, uses real-time PCR (Methylight) to quantify DNA methylation and is likely to add considerably to the volume and quality of data in the public domain.
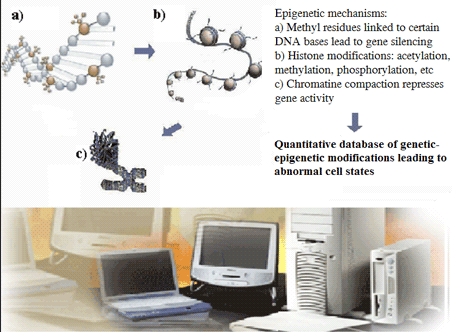
The structure and development of a tool to provide input to a model of genetic-epigenetic interactions, has a number of key requirements. Firstly, the data must be well-organised, validated and readily accessible to the user for immediate comparison and descriptive analysis. Secondly, the context and standard of the data provision for epigenetic information and that of cancer-related fields must be clearly defined. Thirdly, a visualisation capability is clearly desirable for the end-user, together with more sophisticated data mining options.
Consequently, our approach is to build a relational database, with a user-friendly interface accessible from the web. The stored information can ultimately be used in predictive statistical and data-mining tools for disease risk assessment and decision support. The prototype database DBEpigen is stored on a MySQL server in Dublin City University, Ireland. The entity-relational scheme was designed with an innovative software DBDesigner, which automatically generates the code in SQL and also the schema in XML, useful by its facility in describing and exchanging data on the web. The proposed language for writing the interface is PHP, as it offers excellent connectivity to a wide range of databases including MySQL. PHP is distinct from client-side JavaScript as the code is executed on the server, providing a strong tool to write a database-enabled web page.
The database will concentrate on combining dispersed information, with particular focus on storing the causal relations between different entities, such as environmental factors, epigenetic events, genetic events and final cell states. It is intended to store current research data and incorporate refinements as soon as these become available.
Links:
http://student.computing.dcu.ie/phpmyadmin/
Login: guestuser
Password: guestpass
Please contact:
Ana Barat
School of Computing, Dublin City University, Ireland
Tel: +353 1 700 8449
E-mail: abarat![]() computing.dcu.ie
computing.dcu.ie









