by Jean Clairambault, François Fages and Sylvain Soliman
'Temporal Genomics for Tailored Chronotherapeutics' (Tempo), is a European project partly funded by the European Union's FP6-LifeSciHealth programme. The project investigates the possibility of individual cancer therapeutics by genetic profiling of cellular drug processing mechanisms and their circadian rhythms.
The main principle that guides Tempo is the exploitation of individual variations (genetic polymorphism) encountered in biological mechanisms. These govern the cell metabolism of anticancer drugs, and particularly those mechanisms that are dependent on the molecular circadian clock, to adapt time-scheduled drug delivery regimens (eg using programmable pumps) for individual patients. Every single patient will belong, according to his genetic enzymatic profile, to one in a limited number of dynamic classes or clusters - ideally between three and five - to be defined after biological measurements of toxicity obtained by blood and tissue samples. And his or her class or cluster will determine his or her personal drug delivery regimen.
Tempo is a Specifically Targeted Research European Project (STREP) and has been running since October 2006. It has a relatively short duration of three years. It involves the investigation of drug processing mechanisms in several laboratory rodent strains. Both genders are used, with each bearing one of several different types of implantable tumour. The drug processing mechanisms will be studied at regularly spaced time points (four or six) during 24 hour spans, so as to identify the influence of circadian rhythms.
The aim of the project is to optimize the delivery of two anticancer drugs. One of these is already in use in everyday cancer clinics, but has significant toxic side effects. The other is a more recent discovery and belongs to a new class of drugs: the cyclin-dependent kinase inhibitors (CDKIs). Initial investigations into the enzymatic activation and degradation systems of these two drugs, irinotecan (initially isolated from the Chinese tree Camptotheca acuminata) and seliciclib (isolated from starfish oocytes), will be performed on laboratory rodents by looking at the genomic expression, protein concentration and enzymatic activities of the cell mechanisms. Finally, molecular and physiological human descriptive variables will be gathered in patients with the aim of obtaining theoretically optimal drug delivery time schedules for these two drugs separately.
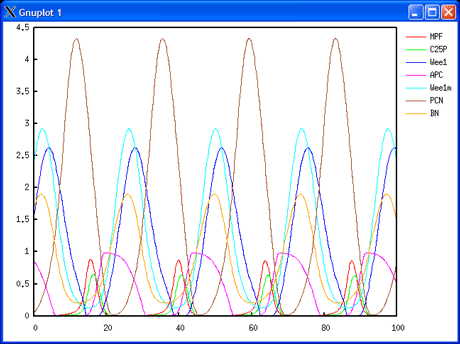
The project includes a work package dedicated to data integration and mathematical modelling, which is managed by INRIA in the research teams Bang and Contraintes at Rocquencourt. Models will be designed both at the individual cell and cell population levels. At the individual cell level, ODE models will be developed in the BIOCHAM environment for molecular systems biology (see http://contraintes.inria.fr/BIOCHAM). These will describe the interrelations between the cell division cycle and the circadian clock, and the pharmacokinetic-pharmocodynamic (PK-PD) mechanisms of drug activity with their genetic polymorphism and circadian variations. At the cell population level, age-structured PDE models of homogeneous cell populations (healthy or tumorous), subject to renewal, will be developed and analysed as targets of cytotoxic drugs. The pharmacological control of the growth of the cell populations under study will be analysed both by a global cell population number and a Malthus exponent for each tissue represented.
Future activities to be developed from this project include the investigation of other anticancer drugs and their synergies, and the definition of optimized chronotherapeutic strategies, individualized for patients in order to minimize unwanted toxic side effects on healthy tissues and to overcome the occurrence of drug resistance.
The Tempo project will be developed in conjunction with other running FP6 European projects to which members of the Tempo consortium already belong, and which are dedicated to modelling of either cancer (M3CSTGT: http://calvino.polito.it/~mcrtn) or drug development (BIOSIM: htttp://biosim.fysik. dtu.dk:8080/biosim/index.jsp).
The project is coordinated by Francis Lévi (INSERM U 776 "Biological Rhythms and Cancers" Paul-Brousse Hospital, Villejuif, Fance). Associated teams of researchers are from fundamental biology and clinical oncology, studying the cell division cycle, pharmacogenomics, or circadian chronobiology at CNRS, INSERM and European universities, with INRIA teams dedicated to mathematical and systems biology, and SMEs accustomed to applied mathematical modelling for pharmaceutical industries and drug delivery device technology.
Link:
http://www.chrono-tempo.org/
Please contact:
Francis Lévi, INSERM, France
Tel: +33 1 45 59 37 20
E-mail: levi-f![]() vjf.inserm.fr
vjf.inserm.fr
Jean Clairambault
INRIA, France
Tel: +33 1 39 63 55 43
E-mail: jean.clairambault![]() inria.fr
inria.fr









