by Kostas Marias, Thanassis Margaritis and Ioannis G. Tollis
In order to assess the clinical importance of models of human pathology (eg cancer), it is necessary to validate them with pre- and post-treatment clinical data. This in turn requires that the size and shape of the tumour, along with structural and physiological information, be determined with high resolution, accuracy and precision. ICS-FORTH has been involved in several research projects addressing image analysis, with the aim of defining optimal methods to robustly extract multiscale anatomical and functional information related to the underlying pathology. This information can be used to initialize and validate models of pathophysiology and to test simulations and predictions of the success of therapeutic regimes.
Imaging techniques in the field of medicine have focused on providing anatomical information, particularly relating to human bones, dense tissue and arteries. PET and functional MRI allow the study of various pathological processes via radio-labelled tracers (PET) or pharmaco-kinetic models in contrast-enhanced (CE) MRI. The whole field of molecular medicine and molecular imaging is opening up new possibilities for targeted assessment of disease and disease mechanisms. In addition, microarray imaging has created exciting possibilities for measuring gene differential expression and defining new disease biomarkers.
In order to improve existing models of human pathophysiology it is essential to robustly extract multiscale information, both anatomical and physiological. For example, consider the importance to cancer modelling of determining genetic profile changes and global changes in tumour size and density, through the analysis of temporal biomedical data. In many cases, common problems arise over different scales (eg geometrical inconsistencies over time). It is therefore important to develop generic tools for multiscale temporal analysis so that time-dependent pathophysiological information can be robustly extracted and visualized. Such information is crucial for initializing, inspiring and validating 4D models of human functions. For example, in the case of in silico models of cancer, 3D voxels should be classified as 'proliferating', 'necrotic', etc.
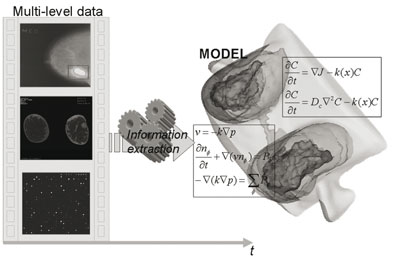
From the imaging standpoint it is essential to stress two points. First, there is a need for a holistic understanding of pathophysiology and this clearly implies a multidisciplinary approach. To this end, molecular and genetic imaging offers unique opportunities to better understand pathophysiology on smaller scales, and to build multilevel models.
Second, it is necessary for the extraction of pathophysiological information to pre-process biomedical data at all possible scales (eg medical images, microarray scans). Thus, multiscale information extraction is necessary to "individualize" a given model. This concept is schematically illustrated in Figure 1, where the extracted multilevel temporal information (eg from microarrays, CE-MRI and mammography) is driven into the corresponding multilevel model of breast cancer.
FORTH has been involved in several aspects of multimodal information extraction (see publication list), and is currently participating in the cancer modelling WP of the ACGT IP project. Some areas of research include:
Extracting regions of interest from biomedical data: Several algorithms have been developed for identifying important structures and features from biomedical images. For example, tumour segmentation is achieved using pharmacokinetic models of gadolinium uptake with contrast-enhanced MRI and microarray spot segmentation through a combination of the two different information channels, ie Cy3 and Cy5.
Geometrical normalization: A number of algorithms have been developed for aligning temporal biomedical data from the same patient, in order to correct geometrical distortions and allow the robust extraction of pathophysiological parameters. Such algorithms can be widely applied, from breast cancer to newer applications such as molecular imaging (eg for correcting time-dependent geometries in 2D molecular optical imaging studies) and microarray imaging (for aligning different channels prior to computing differential expressions).
Intensity normalization: Biomedical measurements often 'mask' true underlying physiological properties due to the image formation process. The nonlinearities introduced by varying imaging conditions may significantly alter the image intensity profile and reduce the efficiency of generic analysis algorithms. This is a well-known problem in cancer imaging (eg mammography). In microarray imaging, several nonlinearities in the experimental process render the measured expression values prone to variability, and often to poor reproducibility.
FORTH ICS and IMBB have co-developed algorithms for normalizing microarray values. This was done by exploiting the fact that the gene expression values (eg from the Cy3 and Cy5 matrices) should ideally follow a linear trend, and considering a "ground truth" subset of imaged genes that is known a priori to be the same in both channels (Cy3 and Cy5).
Visualization: This is a fundamental aspect of biomedical data information fusion that is typically less well addressed in the literature, but which can dramatically increase the clinical utility of a solution if implemented intelligently. To develop patient-specific models it is important to implement appropriate tools for information visualization at all levels, from gene networks to cellular processes and organ function. To this end, FORTH-ICS has developed novel tools and algorithms for visualizing complex biological networks.
Links:
http://www.ics.forth.gr/bmi/publications.jsp
http://www.ics.forth.gr/~kmarias/publications.htm
http://www.eu-acgt.org/
Please contact:
Kostas Marias
ICS-FORTH, Greece
Tel: +30 2810 391672
E-mail: kmarias![]() ics.forth.gr
ics.forth.gr









