by Ioannis Tsamardinos
What gene's expression is causing another one to be expressed? Which combination of mutations is causing disease? Knowledge of causal relations is paramount in simulating the digital patient, understanding the mechanisms of disease, designing drugs and treating patients. Recent theoretical and algorithmic advances in the discovery of causal relations from observational data promise to boost our biomedical knowledge.
Perhaps the most basic scientific tool for advancing knowledge is the randomized controlled experiment, where a quantity A (eg smoking) is manipulated in a controlled manner on a random population and the effects are measured on a quantity B (eg development of lung cancer). A significant portion of classical statistics is concerned with soundly inferring from the measurements of the experiment whether or not A (probabilistically) causes B. Unfortunately, particularly in biomedicine, such experiments are often costly (in terms of both time and money), unethical, or even impossible. Nevertheless, a wealth of observational data is often available to researchers; the issue is then to identify the most useful probable causal hypotheses on which to focus.
The induction of causal relations from observational data has traditionally been an anathema in statistics. Data analysts in bioinformatics commonly state their results in a covered way: "the function of protein A is related to the function of protein B", "our analysis has identified the relevant genes for the expression of gene B". Behind the use of the terms "related" and "relevant" is typically implied more than a statistical association: a causal relation. Correlation is not causation; yet the existence of some correlations and the absence of others can lead us to induce under some fairly broad conditions the existence or absence of certain causal relations. Since the end of the 80s, the work on formal theories of causality and causal induction by Spirtes, Glymour, Pearl, Cooper and others has been gaining ground. In 2003, Clive Granger won the Nobel Prize in Economics for his work on the causal analysis of observational economic time series, giving more respectability to the field. Several articles have already appeared in the bioinformatics literature employing causal techniques, and related conferences and workshops (such as the recent NIPS workshop on causal feature selection) have been held.
Several theories of causality have corresponding graphical means for representing causal relations, such as Structural Equation Models and the more recent Causal Bayesian Networks (see Figure 1). Algorithms exist for inducing such networks from observational data. Our recent work has extended learning Bayesian Networks with tens of thousands of variables and unprecedented accuracy. In addition, a growing body of our work is focused on causal variable selection: when the time available or the number of variables does not permit the construction of the full Bayesian Network, algorithms can identify the local causal neighbourhood of a variable of interest in reasonable time, for example, the causes and effects of the expression levels of a gene. The causal local neighbourhood (related to the concept of the Markov Blanket) is under some conditions the smallest variable subset required for optimal prediction. Our algorithms often outperform traditional non-causal variable selection algorithms for prediction, and in addition the selected variables have known causal relations to the target. Our group, along with other researchers, is carrying out theoretical work to accompany this algorithmic work. This includes looking at the conditions under which the relations found correspond to causal relations, lifting or changing the set of assumptions for causal discovery and extending it to different types of data. Algorithms now exist for identifying hidden (unobserved) variables that cause (change the distribution of) the variable of interest, that explicitly model selection bias, model feedback loops and other interesting situations.
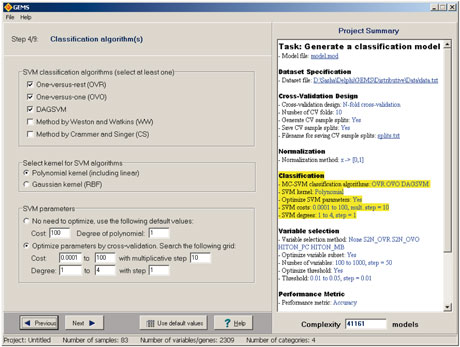
We have developed a couple of tools for applying causal data-mining techniques to real data. The first is Causal Explorer, a library of algorithms for learning Bayesian Networks and identifying the causal neighbourhood of a target variable. The second is the Gene Expression Model Selector or GEMS, which automates the mining of gene expression data with the option of using some of the causal methods mentioned above.
In moving to the Institute of Computer Science at the Foundation for Research and Technology, Hellas and the Biomedical Informatics Laboratory, I will be extending this line of work in several dimensions. In our plans are new algorithms, more theoretical results, and enhanced tools for inducing and mining causality; in addition, the application of such methods to biomedical data to answer specific biological questions. In particular, for the Digital Patient, our methods could identify from data the factors that need to be modeled in order to simulate the development of a disease or a human subsystem malfunction.
Please contact:
Ioannis Tsamardinos
ICS-FORTH, Greece
Tel: +30 2810 391 617
E-mail: tsamard![]() ics.forth.gr
ics.forth.gr









